Scaling by shrinking: empowering single-cell 'omics' with microfluidic devices
- PMID: 28392571
- PMCID: PMC5495114
- DOI: 10.1038/nrg.2017.15
Scaling by shrinking: empowering single-cell 'omics' with microfluidic devices
Abstract
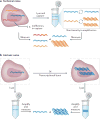
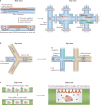
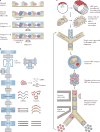
Recent advances in cellular profiling have demonstrated substantial heterogeneity in the behaviour of cells once deemed 'identical', challenging fundamental notions of cell 'type' and 'state'. Not surprisingly, these findings have elicited substantial interest in deeply characterizing the diversity, interrelationships and plasticity among cellular phenotypes. To explore these questions, experimental platforms are needed that can extensively and controllably profile many individual cells. Here, microfluidic structures - whether valve-, droplet- or nanowell-based - have an important role because they can facilitate easy capture and processing of single cells and their components, reducing labour and costs relative to conventional plate-based methods while also improving consistency. In this article, we review the current state-of-the-art methodologies with respect to microfluidics for mammalian single-cell 'omics' and discuss challenges and future opportunities.
Conflict of interest statement
The authors declare competing interests: see
Figures


References
-
- Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. A demonstration of how covariation in gene expression in scRNA-Seq data can be used to identify cellular circuits and molecular drivers of behaviour in seemingly identical immune cells. It also nicely demonstrates the use of microfluidic devices for targeted single-cell mRNA measurements (STA). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

