Anti-Biofilm Activity of a Self-Aggregating Peptide against Streptococcus mutans
- PMID: 28392782
- PMCID: PMC5364132
- DOI: 10.3389/fmicb.2017.00488
Anti-Biofilm Activity of a Self-Aggregating Peptide against Streptococcus mutans
Abstract
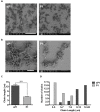
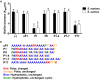
Streptococcus mutans is the primary agent of dental cavities, in large part due to its ability to adhere to teeth and create a molecular scaffold of glucan polysaccharides on the tooth surface. Disrupting the architecture of S. mutans biofilms could help undermine the establishment of biofilm communities that cause cavities and tooth decay. Here we present a synthetic peptide P1, derived from a tick antifreeze protein, which significantly reduces S. mutans biofilm formation. Incubating cells with this peptide decreased biofilm biomass by approximately 75% in both a crystal violet microplate assay and an in vitro tooth model using saliva-coated hydroxyapatite discs. Bacteria treated with peptide P1 formed irregular biofilms with disconnected aggregates of cells and exopolymeric matrix that readily detached from surfaces. Peptide P1 can bind directly to S. mutans cells but does not possess bactericidal activity. Anti-biofilm activity was correlated with peptide aggregation and β-sheet formation in solution, and alternative synthetic peptides of different lengths or charge distribution did not inhibit biofilms. This anti-biofilm peptide interferes with S. mutans biofilm formation and architecture, and may have future applications in preventing bacterial buildup on teeth.
Keywords: Gram-positive cocci; Streptococcus mutans; aggregation; anti-biofilm; biofilm; synthetic peptide.
Figures






References
-
- Basak S., Rajurkar M., Attal R., Mallick S. (2013). “Biofilms: a challenge to medical fraternity in infection control,” in Infection Control ed. Basak S. (Rijeka: InTech; ), 57–74. 10.5772/56704 - DOI
-
- Besingi R. N., Wenderska I. B., Senadheera D. B., Cvitkovitch D. G., Long J. R., Wen Z. T., et al. (2017). Functional Amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c. Microbiology 10.1099/mic.0.000443 [Epub ahead of print]. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

