Global gene expression in Escherichia coli, isolated from the diseased ocular surface of the human eye with a potential to form biofilm
- PMID: 28392838
- PMCID: PMC5379667
- DOI: 10.1186/s13099-017-0164-2
Global gene expression in Escherichia coli, isolated from the diseased ocular surface of the human eye with a potential to form biofilm
Abstract
Background: Escherichia coli, the gastrointestinal commensal, is also known to cause ocular infections such as conjunctivitis, keratitis and endophthalmitis. These infections are normally resolved by topical application of an appropriate antibiotic. But, at times these E. coli are resistant to the antibiotic and this could be due to formation of a biofilm. In this study ocular E. coli from patients with conjunctivitis, keratitis or endophthalmitis were screened for their antibiotic susceptibility and biofilm formation potential. In addition DNA-microarray analysis was done to identify genes that are involved in biofilm formation and antibiotic resistance.
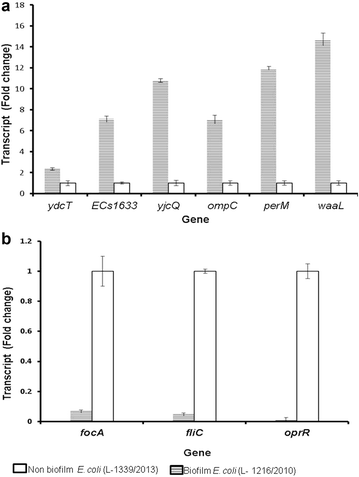
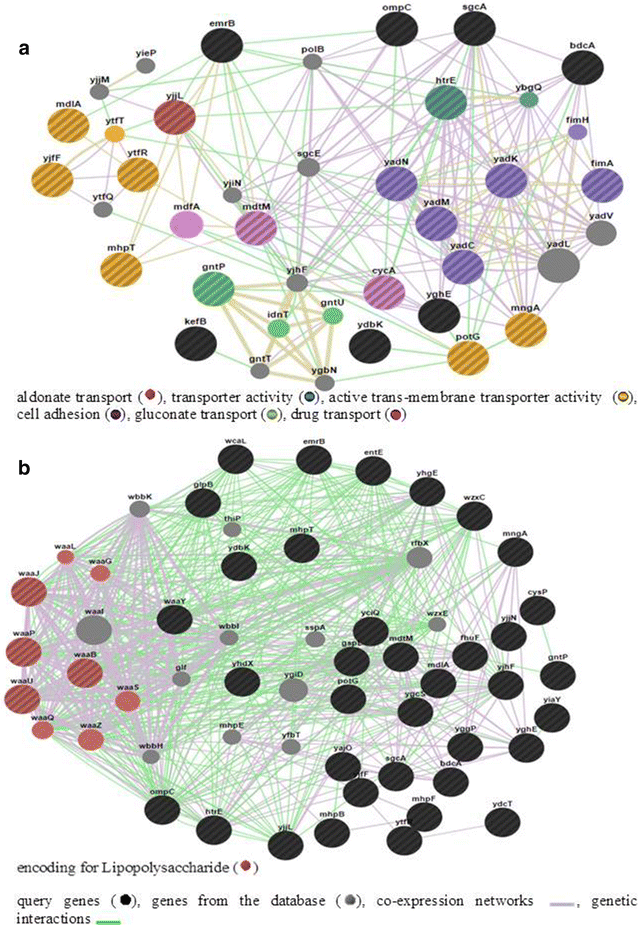
Results: Out of 12 ocular E. coli isolated from patients ten isolates were resistant to one or more of the nine antibiotics tested and majority of the isolates were positive for biofilm formation. In E. coli L-1216/2010, the best biofilm forming isolate, biofilm formation was confirmed by scanning electron microscopy. Confocal laser scanning microscopic studies indicated that the thickness of the biofilm increased up to 72 h of growth. Further, in the biofilm phase, E. coli L-1216/2010 was 100 times more resistant to the eight antibiotics tested compared to planktonic phase. DNA microarray analysis indicated that in biofilm forming E. coli L-1216/2010 genes encoding biofilm formation such as cell adhesion genes, LPS production genes, genes required for biofilm architecture and extracellular matrix remodeling and genes encoding for proteins that are integral to the cell membrane and those that influence antigen presentation are up regulated during biofilm formation. In addition genes that confer antimicrobial resistance such as genes encoding antimicrobial efflux (mdtM and cycA), virulence (insQ, yjgK), toxin production (sat, yjgK, chpS, chpB and ygjN), transport of amino-acids and other metabolites (cbrB, cbrC, hisI and mglB) are also up regulated. These genes could serve as potential targets for developing strategies for hacking biofilms and overcoming antibiotic resistance.
Conclusions: This is the first study on global gene expression in antibiotic resistant ocular E. coli with a potential to form biofilm. Using native ocular isolates for antibiotic susceptibility testing, for biofilm formation and global gene expression is relevant and more acceptable than using type strains or non clinical strains which do not necessarily mimic the native isolate.
Keywords: Antibiotic susceptibility; Biofilm; DNA microarray; Differential expression of genes; Ocular E. coli.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

