Baculovirus LEF-11 Hijack Host ATPase ATAD3A to Promote Virus Multiplication in Bombyx mori cells
- PMID: 28393927
- PMCID: PMC5385504
- DOI: 10.1038/srep46187
Baculovirus LEF-11 Hijack Host ATPase ATAD3A to Promote Virus Multiplication in Bombyx mori cells
Abstract
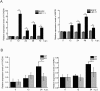
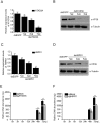
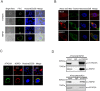
Research on molecular mechanisms that viruses use to regulate the host apparatus is important in virus infection control and antiviral therapy exploration. Our previous research showed that the Bombyx mori nucleopolyhedrovirus (BmNPV) LEF-11 localized to dense regions of the cell nucleus and is required for viral DNA replication. Herein, we examined the mechanism of LEF-11 on BmNPV multiplication and demonstrated that baculovirus LEF-11 interacts with Bombyx mori ATAD3A and HSPD1 (HSP60) protein. Furthermore, we showed that LEF-11 has the ability to induce and up-regulate the expression of ATAD3A and HSPD1, phenomena that were both reversed upon knockdown of lef-11. Our findings showed that ATAD3A and HSPD1 were necessary and contributed to BmNPV multiplication in Bombyx mori cells. Moreover, ATAD3A was found to directly interact with HSPD1. Interestingly, ATAD3A was required for the expression of HSPD1, while the knockdown of HSPD1 had no obvious effect on the expression level of ATAD3A. Taken together, the data presented in the current study demonstrated that baculovirus LEF-11 hijacks the host ATPase family members, ATAD3A and HSPD1, efficiently promote the multiplication of the virus. This study furthers our understanding of how baculovirus modulates energy metabolism of the host and provides a new insight into the molecular mechanisms of antiviral research.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Bombyx mori nucleopolyhedrovirus LEF-2 disrupts the cell cycle in the G2/M phase by triggering a host cell DNA damage response.Insect Mol Biol. 2025 Feb;34(1):81-93. doi: 10.1111/imb.12951. Epub 2024 Aug 16. Insect Mol Biol. 2025. PMID: 39150688
-
Functional characterization of Bombyx mori nucleopolyhedrovirus mutant lacking late expression factor 9.Acta Virol. 2016;60(3):281-9. doi: 10.4149/av_2016_03_281. Acta Virol. 2016. PMID: 27640438
-
Comprehensive Profiling of Lysine Acetylome in Baculovirus Infected Silkworm (Bombyx mori) Cells.Proteomics. 2018 Jan;18(1). doi: 10.1002/pmic.201700133. Proteomics. 2018. PMID: 29150924
-
The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori.Insect Biochem Mol Biol. 2014 May;48:1-7. doi: 10.1016/j.ibmb.2014.02.003. Epub 2014 Feb 20. Insect Biochem Mol Biol. 2014. PMID: 24561307 Review.
-
MicroRNA-Mediated Host-Pathogen Interactions Between Bombyx mori and Viruses.Front Physiol. 2021 May 7;12:672205. doi: 10.3389/fphys.2021.672205. eCollection 2021. Front Physiol. 2021. PMID: 34025458 Free PMC article. Review.
Cited by
-
Host AAA+ ATPase TER94 Plays Critical Roles in Building the Baculovirus Viral Replication Factory and Virion Morphogenesis.J Virol. 2020 Feb 28;94(6):e01674-19. doi: 10.1128/JVI.01674-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896597 Free PMC article.
-
Protein-Gene Orthology in Baculoviridae: An Exhaustive Analysis to Redefine the Ancestrally Common Coding Sequences.Viruses. 2023 Apr 29;15(5):1091. doi: 10.3390/v15051091. Viruses. 2023. PMID: 37243176 Free PMC article.
-
Baculoviruses manipulate host lipid metabolism via adipokinetic hormone signaling to induce climbing behavior.PLoS Pathog. 2025 Jan 31;21(1):e1012932. doi: 10.1371/journal.ppat.1012932. eCollection 2025 Jan. PLoS Pathog. 2025. PMID: 39888969 Free PMC article.
-
Regulation of Bombyx mori-BmNPV Protein Interactions: Study Strategies and Molecular Mechanisms.Viruses. 2025 Jul 20;17(7):1017. doi: 10.3390/v17071017. Viruses. 2025. PMID: 40733633 Free PMC article. Review.
-
Excision of Nucleopolyhedrovirus Form Transgenic Silkworm Using the CRISPR/Cas9 System.Front Microbiol. 2018 Feb 16;9:209. doi: 10.3389/fmicb.2018.00209. eCollection 2018. Front Microbiol. 2018. PMID: 29503634 Free PMC article.
References
-
- Berenguer M. Outcome of posttransplantation hepatitis C virus disease–is it the host, the virus, or how we modify the host and/or the virus? Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society 8, 889–891, doi: 10.1053/jlts.2002.35841 (2002). - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

