Natural non-homologous recombination led to the emergence of a duplicated V3-NS5A region in HCV-1b strains associated with hepatocellular carcinoma
- PMID: 28394908
- PMCID: PMC5386276
- DOI: 10.1371/journal.pone.0174651
Natural non-homologous recombination led to the emergence of a duplicated V3-NS5A region in HCV-1b strains associated with hepatocellular carcinoma
Abstract
Background: The emergence of new strains in RNA viruses is mainly due to mutations or intra and inter-genotype homologous recombination. Non-homologous recombinations may be deleterious and are rarely detected. In previous studies, we identified HCV-1b strains bearing two tandemly repeated V3 regions in the NS5A gene without ORF disruption. This polymorphism may be associated with an unfavorable course of liver disease and possibly involved in liver carcinogenesis. Here we aimed at characterizing the origin of these mutant strains and identifying the evolutionary mechanism on which the V3 duplication relies.
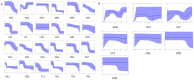
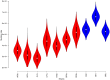
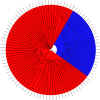
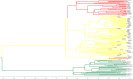
Methods: Direct sequencing of the entire NS5A and E1 genes was performed on 27 mutant strains. Quasispecies analyses in consecutive samples were also performed by cloning and sequencing the NS5A gene for all mutant and wild strains. We analyzed the mutant and wild-type sequence polymorphisms using Bayesian methods to infer the evolutionary history of and the molecular mechanism leading to the duplication-like event.
Results: Quasispecies were entirely composed of exclusively mutant or wild-type strains respectively. Mutant quasispecies were found to have been present since contamination and had persisted for at least 10 years. This V3 duplication-like event appears to have resulted from non-homologous recombination between HCV-1b wild-type strains around 100 years ago. The association between increased liver disease severity and these HCV-1b mutants may explain their persistence in chronically infected patients.
Conclusions: These results emphasize the possible consequences of non-homologous recombination in the emergence and severity of new viral diseases.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

