DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning
- PMID: 28395661
- PMCID: PMC5387360
- DOI: 10.1186/s13059-017-1189-z
DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning
Erratum in
-
Erratum to: DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning.Genome Biol. 2017 May 12;18(1):90. doi: 10.1186/s13059-017-1233-z. Genome Biol. 2017. PMID: 28499443 Free PMC article. No abstract available.
Abstract
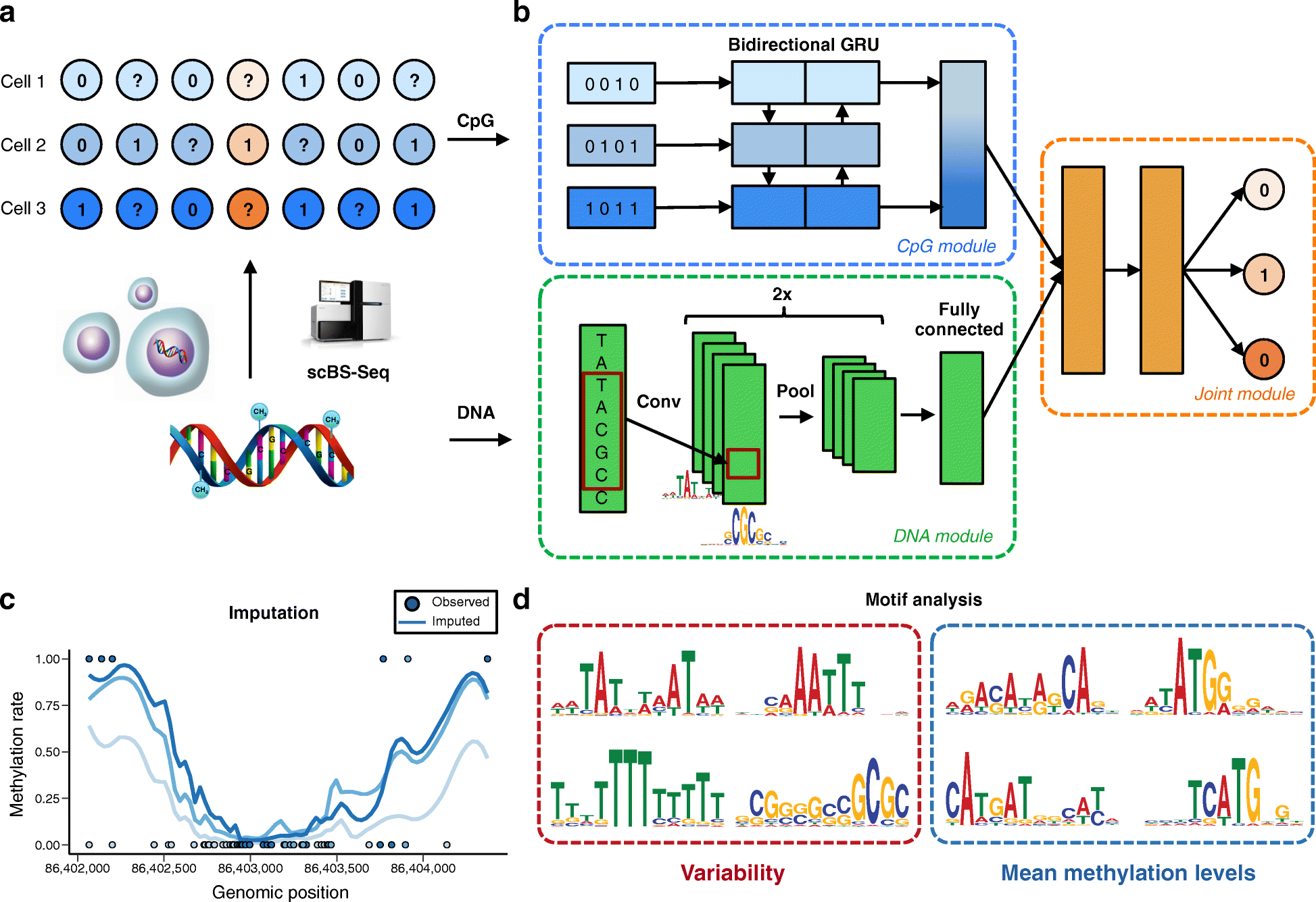
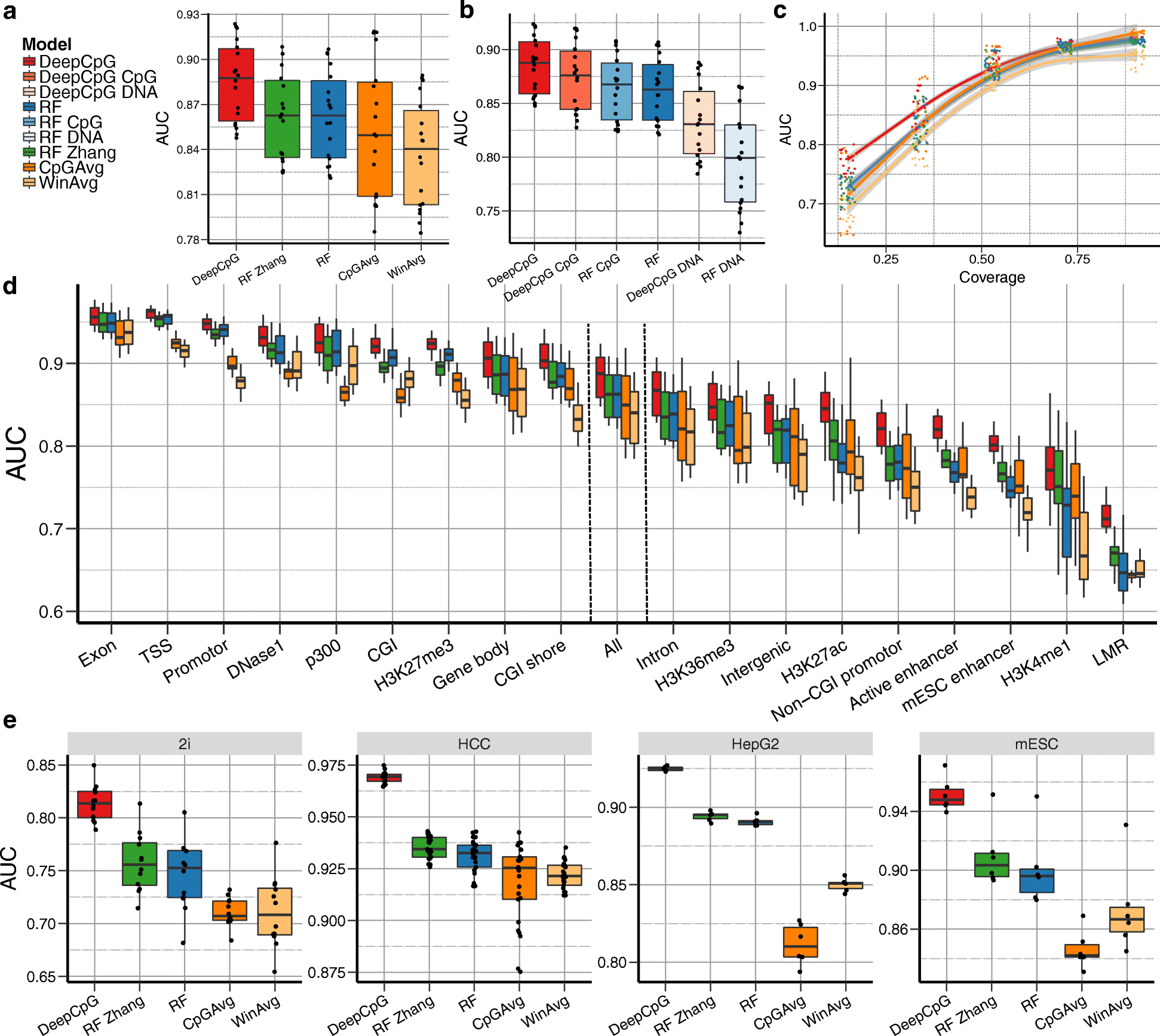
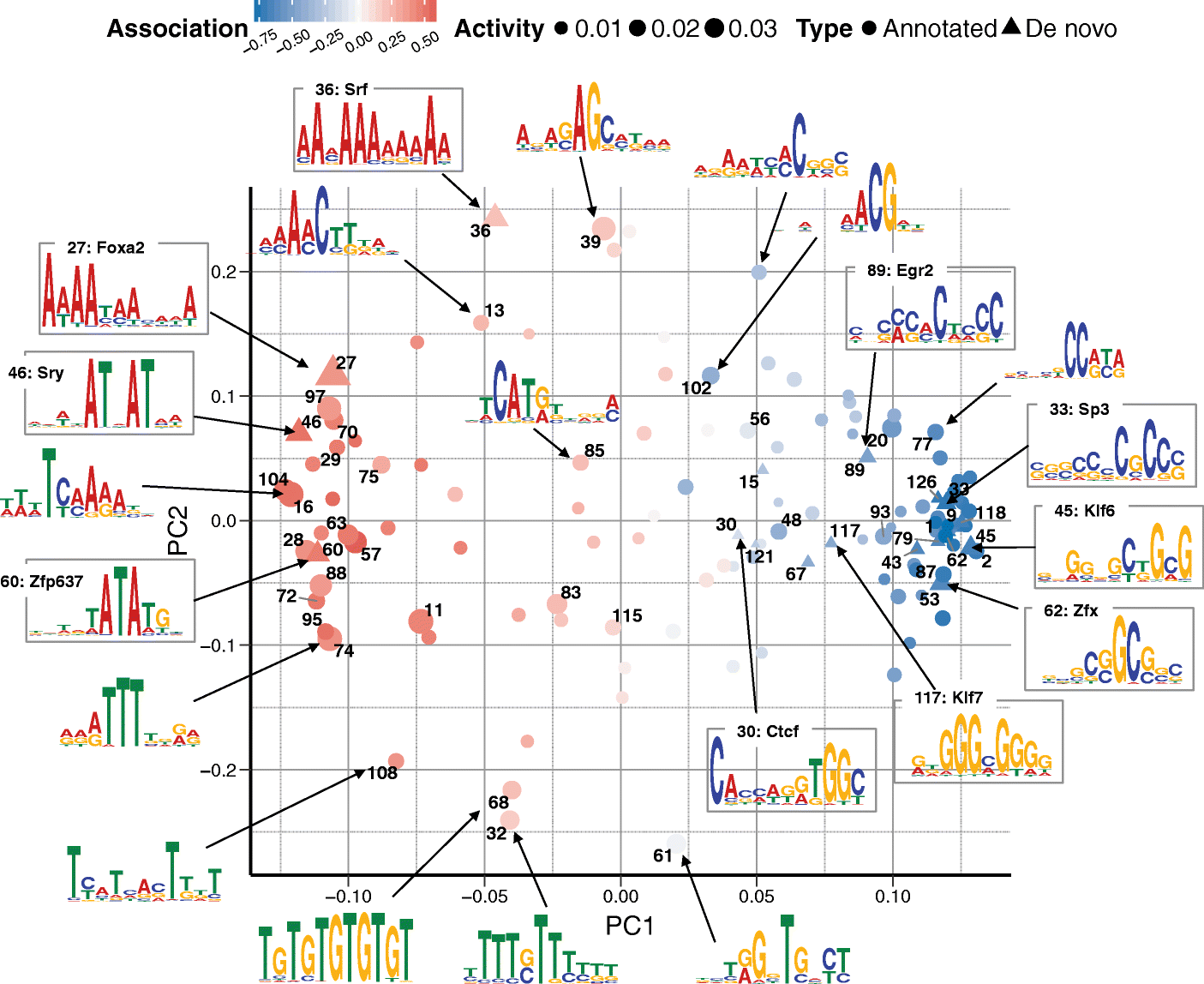
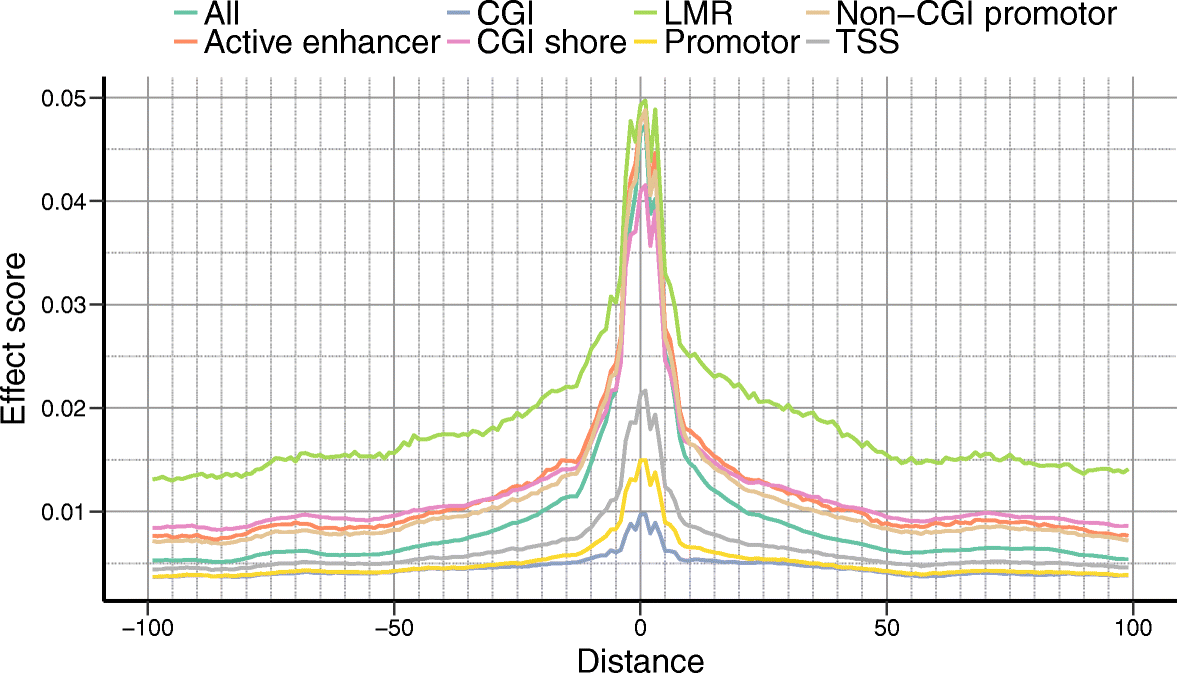
Recent technological advances have enabled DNA methylation to be assayed at single-cell resolution. However, current protocols are limited by incomplete CpG coverage and hence methods to predict missing methylation states are critical to enable genome-wide analyses. We report DeepCpG, a computational approach based on deep neural networks to predict methylation states in single cells. We evaluate DeepCpG on single-cell methylation data from five cell types generated using alternative sequencing protocols. DeepCpG yields substantially more accurate predictions than previous methods. Additionally, we show that the model parameters can be interpreted, thereby providing insights into how sequence composition affects methylation variability.
Keywords: Artificial neural network; DNA methylation; Deep learning; Epigenetics; Machine learning; Single-cell genomics.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

