Elucidation of the anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins
- PMID: 28395732
- PMCID: PMC5388539
- DOI: 10.7554/eLife.23905
Elucidation of the anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins
Abstract
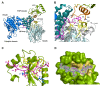
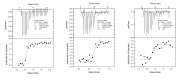
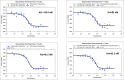
Autophagy is a conserved cellular process involved in the elimination of proteins and organelles. It is also used to combat infection with pathogenic microbes. The intracellular pathogen Legionella pneumophila manipulates autophagy by delivering the effector protein RavZ to deconjugate Atg8/LC3 proteins coupled to phosphatidylethanolamine (PE) on autophagosomal membranes. To understand how RavZ recognizes and deconjugates LC3-PE, we prepared semisynthetic LC3 proteins and elucidated the structures of the RavZ:LC3 interaction. Semisynthetic LC3 proteins allowed the analysis of structure-function relationships. RavZ extracts LC3-PE from the membrane before deconjugation. RavZ initially recognizes the LC3 molecule on membranes via its N-terminal LC3-interacting region (LIR) motif. The RavZ α3 helix is involved in extraction of the PE moiety and docking of the acyl chains into the lipid-binding site of RavZ that is related in structure to that of the phospholipid transfer protein Sec14. Thus, Legionella has evolved a novel mechanism to specifically evade host autophagy.
Keywords: GDI; Legionella pneumophila; Sec14; autophagy; biochemistry; biophysics; host-pathogen interaction; native chemical ligation; none; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures










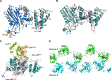




Similar articles
-
Lift and cut: Anti-host autophagy mechanism of Legionella pneumophila.Autophagy. 2017 Aug 3;13(8):1467-1469. doi: 10.1080/15548627.2017.1327943. Epub 2017 Jun 9. Autophagy. 2017. PMID: 28598235 Free PMC article.
-
Distinct Mechanisms for Processing Autophagy Protein LC3-PE by RavZ and ATG4B.Chembiochem. 2020 Dec 1;21(23):3377-3382. doi: 10.1002/cbic.202000359. Epub 2020 Aug 25. Chembiochem. 2020. PMID: 32686895 Free PMC article.
-
LIR motifs and the membrane-targeting domain are complementary in the function of RavZ.BMB Rep. 2019 Dec;52(12):700-705. doi: 10.5483/BMBRep.2019.52.12.211. BMB Rep. 2019. PMID: 31722778 Free PMC article.
-
A Structural View of Xenophagy, a Battle between Host and Microbes.Mol Cells. 2018 Jan 31;41(1):27-34. doi: 10.14348/molcells.2018.2274. Epub 2018 Jan 23. Mol Cells. 2018. PMID: 29370690 Free PMC article. Review.
-
Autophagy Evasion and Endoplasmic Reticulum Subversion: The Yin and Yang of Legionella Intracellular Infection.Annu Rev Microbiol. 2016 Sep 8;70:413-33. doi: 10.1146/annurev-micro-102215-095557. Annu Rev Microbiol. 2016. PMID: 27607556 Review.
Cited by
-
3,4‑Dihydroxyacetophenone attenuates oxidative stress‑induced damage to HUVECs via regulation of the Nrf2/HO‑1 pathway.Mol Med Rep. 2022 Jun;25(6):199. doi: 10.3892/mmr.2022.12715. Epub 2022 Apr 27. Mol Med Rep. 2022. PMID: 35475506 Free PMC article.
-
How to rewire the host cell: A home improvement guide for intracellular bacteria.J Cell Biol. 2017 Dec 4;216(12):3931-3948. doi: 10.1083/jcb.201701095. Epub 2017 Nov 2. J Cell Biol. 2017. PMID: 29097627 Free PMC article. Review.
-
Differential roles of N- and C-terminal LIR motifs in the catalytic activity and membrane targeting of RavZ and ATG4B proteins.BMB Rep. 2024 Nov;57(11):497-502. doi: 10.5483/BMBRep.2024-0084. BMB Rep. 2024. PMID: 39384175 Free PMC article.
-
A cancer associated somatic mutation in LC3B attenuates its binding to E1-like ATG7 protein and subsequent lipidation.Autophagy. 2019 Mar;15(3):438-452. doi: 10.1080/15548627.2018.1525476. Epub 2018 Oct 8. Autophagy. 2019. PMID: 30238850 Free PMC article.
-
Development of selective deconjugases for membrane-anchored LC3A/B in post-mitotic neurons.Mol Brain. 2025 Feb 12;18(1):11. doi: 10.1186/s13041-025-01184-z. Mol Brain. 2025. PMID: 39940005 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity : all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

