The impact of stratified immunity on the transmission dynamics of influenza
- PMID: 28395850
- PMCID: PMC5628170
- DOI: 10.1016/j.epidem.2017.03.003
The impact of stratified immunity on the transmission dynamics of influenza
Abstract
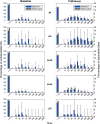
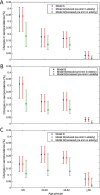
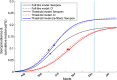
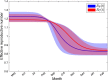
Although empirical studies show that protection against influenza infection in humans is closely related to antibody titres, influenza epidemics are often described under the assumption that individuals are either susceptible or not. Here we develop a model in which antibody titre classes are enumerated explicitly and mapped onto a variable scale of susceptibility in different age groups. Fitting only with pre- and post-wave serological data during 2009 pandemic in Hong Kong, we demonstrate that with stratified immunity, the timing and the magnitude of the epidemic dynamics can be reconstructed more accurately than is possible with binary seropositivity data. We also show that increased infectiousness of children relative to adults and age-specific mixing are required to reproduce age-specific seroprevalence observed in Hong Kong, while pre-existing immunity in the elderly is not. Overall, our results suggest that stratified immunity in an aged-structured heterogeneous population plays a significant role in determining the shape of influenza epidemics.
Keywords: Age-specific seroprevalence; Antibody responses; Epidemic model; Inferring transmission dynamics; Influenza; Stratified immunity.
Copyright © 2017. Published by Elsevier B.V.
Figures

References
-
- Achonu C., Rosella L., Gubbay J.B., Deeks S., Rebbapragada A., Mazzulli T., Willison D., Foisy J., McGeer A., Johnson I., LaFreniere M., Johnson C., Willmore J., Yue C., Crowcroft N.S. Seroprevalence of pandemic influenza H1N1 in Ontario from January 2009–May 2010. PLoS ONE. 2011;6(11) - PMC - PubMed
-
- Allen L.J.S., Lahodny G.E. Extinction thresholds in deterministic and stochastic epidemic models. J. Biol. Dyn. 2012;6(2):590–611. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

