Association of the Cold Shock DEAD-Box RNA Helicase RhlE to the RNA Degradosome in Caulobacter crescentus
- PMID: 28396352
- PMCID: PMC5472812
- DOI: 10.1128/JB.00135-17
Association of the Cold Shock DEAD-Box RNA Helicase RhlE to the RNA Degradosome in Caulobacter crescentus
Abstract
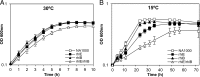
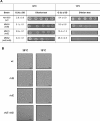
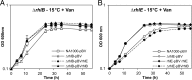
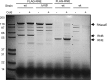
In diverse bacterial lineages, multienzyme assemblies have evolved that are central elements of RNA metabolism and RNA-mediated regulation. The aquatic Gram-negative bacterium Caulobacter crescentus, which has been a model system for studying the bacterial cell cycle, has an RNA degradosome assembly that is formed by the endoribonuclease RNase E and includes the DEAD-box RNA helicase RhlB. Immunoprecipitations of extracts from cells expressing an epitope-tagged RNase E reveal that RhlE, another member of the DEAD-box helicase family, associates with the degradosome at temperatures below those optimum for growth. Phenotype analyses of rhlE, rhlB, and rhlE rhlB mutant strains show that RhlE is important for cell fitness at low temperature and its role may not be substituted by RhlB. Transcriptional and translational fusions of rhlE to the lacZ reporter gene and immunoblot analysis of an epitope-tagged RhlE indicate that its expression is induced upon temperature decrease, mainly through posttranscriptional regulation. RNase E pulldown assays show that other proteins, including the transcription termination factor Rho, a second DEAD-box RNA helicase, and ribosomal protein S1, also associate with the degradosome at low temperature. The results suggest that the RNA degradosome assembly can be remodeled with environmental change to alter its repertoire of helicases and other accessory proteins.IMPORTANCE DEAD-box RNA helicases are often present in the RNA degradosome complex, helping unwind secondary structures to facilitate degradation. Caulobacter crescentus is an interesting organism to investigate degradosome remodeling with change in temperature, because it thrives in freshwater bodies and withstands low temperature. In this study, we show that at low temperature, the cold-induced DEAD-box RNA helicase RhlE is recruited to the RNA degradosome, along with other helicases and the Rho protein. RhlE is essential for bacterial fitness at low temperature, and its function may not be complemented by RhlB, although RhlE is able to complement for rhlB loss. These results suggest that RhlE has a specific role in the degradosome at low temperature, potentially improving adaptation to this condition.
Keywords: Caulobacter crescentus; DEAD-box RNA helicase; RNA degradosome; RhlE; cold shock.
Copyright © 2017 Aguirre et al.
Figures


References
-
- Gorbalenya AE, Koonin EV. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol 3:419–429. doi:10.1016/S0959-440X(05)80116-2. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

