Correlation of Meiotic DSB Formation and Transcription Initiation Around Fission Yeast Recombination Hotspots
- PMID: 28396503
- PMCID: PMC5499187
- DOI: 10.1534/genetics.116.197954
Correlation of Meiotic DSB Formation and Transcription Initiation Around Fission Yeast Recombination Hotspots
Abstract
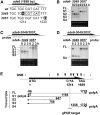
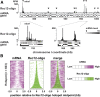
Meiotic homologous recombination, a critical event for ensuring faithful chromosome segregation and creating genetic diversity, is initiated by programmed DNA double-strand breaks (DSBs) formed at recombination hotspots. Meiotic DSB formation is likely to be influenced by other DNA-templated processes including transcription, but how DSB formation and transcription interact with each other has not been understood well. In this study, we used fission yeast to investigate a possible interplay of these two events. A group of hotspots in fission yeast are associated with sequences similar to the cyclic AMP response element and activated by the ATF/CREB family transcription factor dimer Atf1-Pcr1. We first focused on one of those hotspots, ade6-3049, and Atf1. Our results showed that multiple transcripts, shorter than the ade6 full-length messenger RNA, emanate from a region surrounding the ade6-3049 hotspot. Interestingly, we found that the previously known recombination-activation region of Atf1 is also a transactivation domain, whose deletion affected DSB formation and short transcript production at ade6-3049 These results point to a possibility that the two events may be related to each other at ade6-3049 In fact, comparison of published maps of meiotic transcripts and hotspots suggested that hotspots are very often located close to meiotically transcribed regions. These observations therefore propose that meiotic DSB formation in fission yeast may be connected to transcription of surrounding regions.
Keywords: DNA double-strand break formation; chromatin; meiosis; meiotic recombination; transcription.
Copyright © 2017 by the Genetics Society of America.
Figures




References
-
- Borde V., de Massy B., 2013. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 23: 147–155. - PubMed
-
- Borde V., Goldman A. S., Lichten M., 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

