An mRNA Capping Enzyme Targets FACT to the Active Gene To Enhance the Engagement of RNA Polymerase II into Transcriptional Elongation
- PMID: 28396559
- PMCID: PMC5472824
- DOI: 10.1128/MCB.00029-17
An mRNA Capping Enzyme Targets FACT to the Active Gene To Enhance the Engagement of RNA Polymerase II into Transcriptional Elongation
Abstract
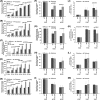
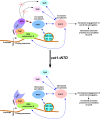
We have recently demonstrated that an mRNA capping enzyme, Cet1, impairs promoter-proximal accumulation/pausing of RNA polymerase II (Pol II) independently of its capping activity in Saccharomyces cerevisiae to control transcription. However, it is still unknown how Pol II pausing is regulated by Cet1. Here, we show that Cet1's N-terminal domain (NTD) promotes the recruitment of FACT (
Keywords: Cet1; FACT; RNA polymerase II; mRNA capping; transcription.
Copyright © 2017 American Society for Microbiology.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
