Human PrimPol activity is enhanced by RPA
- PMID: 28396594
- PMCID: PMC5429719
- DOI: 10.1038/s41598-017-00958-3
Human PrimPol activity is enhanced by RPA
Abstract
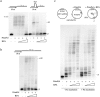
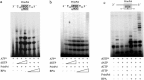
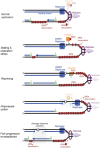
Human PrimPol is a primase belonging to the AEP superfamily with the unique ability to synthesize DNA primers de novo, and a non-processive DNA polymerase able to bypass certain DNA lesions. PrimPol facilitates both mitochondrial and nuclear replication fork progression either acting as a conventional TLS polymerase, or repriming downstream of blocking lesions. In vivo assays have shown that PrimPol is rapidly recruited to sites of DNA damage by interaction with the human replication protein A (RPA). In agreement with previous findings, we show here that the higher affinity of RPA for ssDNA inhibits PrimPol activities in short ssDNA templates. In contrast, once the amount of ssDNA increases up to a length in which both proteins can simultaneously bind ssDNA, as expected during replicative stress conditions, PrimPol and RPA functionally interact, and their binding capacities are mutually enhanced. When using M13 ssDNA as template, RPA stimulated both the primase and polymerase activities of PrimPol, either alone or in synergy with Polε. These new findings supports the existence of a functional PrimPol/RPA association that allows repriming at the exposed ssDNA regions formed in the leading strand upon replicase stalling.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Lipps, G. The replication protein of the Sulfolobus islandicus plasmid pRN1. Biochem Soc Trans32, 240–244, doi:10.1042/ (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

