Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using Biacore™ T200
- PMID: 28396838
- PMCID: PMC5377415
- DOI: 10.1002/2211-5463.12201
Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using Biacore™ T200
Abstract
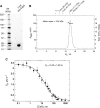
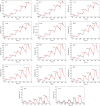
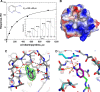
We have established a refined methodology for generating surface plasmon resonance sensor surfaces of recombinant his-tagged human cyclophilin-A. Our orientation-specific stabilisation approach captures his-tagged protein under 'physiological conditions' (150 mm NaCl, pH 7.5) and covalently stabilises it on Ni2+-nitrilotriacetic acid surfaces, very briefly activated for primary amine-coupling reactions, producing very stable and active surfaces (≥ 95% specific activity) of cyclophilin-A. Variation in protein concentration with the same contact time allows straightforward generation of variable density surfaces, with essentially no loss of activity, making the protocol easily adaptable for studying numerous interactions; from very small fragments, ~ 100 Da, to large protein ligands. This new method results in an increased stability and activity of the immobilised protein and allowed us to expand the thermo-kinetic analysis space, and to determine accurate and robust thermodynamic parameters for the cyclophilin-A-cyclosporin-A interaction. Furthermore, the increased sensitivity of the surface allowed identification of a new nonpeptide inhibitor of cyclophilin-A, from a screen of a fragment library. This fragment, 2,3-diaminopyridine, bound specifically with a mean affinity of 248 ± 60 μm. The X-ray structure of this 109-Da fragment bound in the active site of cyclophilin-A was solved to a resolution of 1.25 Å (PDB: 5LUD), providing new insight into the molecular details for a potential new series of nonpeptide cyclophilin-A inhibitors.
Keywords: cyclophilin‐A; inhibitor; nonpeptide; surface plasmon resonance; thermodynamics.
Figures


Similar articles
-
A surface plasmon resonance-based assay for small molecule inhibitors of human cyclophilin A.Anal Biochem. 2005 Oct 15;345(2):214-26. doi: 10.1016/j.ab.2005.06.037. Anal Biochem. 2005. PMID: 16102717 Free PMC article.
-
Covalent affixation of histidine-tagged proteins tethered onto Ni-nitrilotriacetic acid sensors for enhanced surface plasmon resonance detection of small molecule drugs and kinetic studies of antibody/antigen interactions.Analyst. 2019 Jan 14;144(2):587-593. doi: 10.1039/c8an01794h. Analyst. 2019. PMID: 30427328
-
Development and characterization of Ni-NTA-bearing microspheres.Cytometry. 2002 Jul 1;48(3):136-45. doi: 10.1002/cyto.10124. Cytometry. 2002. PMID: 12116359
-
Thermodynamic, kinetic, and structural parameterization of human carbonic anhydrase interactions toward enhanced inhibitor design.Q Rev Biophys. 2018 Jan;51:e10. doi: 10.1017/S0033583518000082. Q Rev Biophys. 2018. PMID: 30912486 Review.
-
Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery.Biochim Biophys Acta. 2014 Jan;1838(1 Pt A):43-55. doi: 10.1016/j.bbamem.2013.04.028. Epub 2013 May 9. Biochim Biophys Acta. 2014. PMID: 23665295 Review.
Cited by
-
Optimization of Cyclophilin B-Targeted Tri-vector Inhibitors for Novel MASH Treatments.J Med Chem. 2025 Mar 27;68(6):6815-6831. doi: 10.1021/acs.jmedchem.5c00301. Epub 2025 Mar 12. J Med Chem. 2025. PMID: 40074291 Free PMC article.
-
Kinetic and structural studies of Trypanosoma and Leishmania phosphofructokinases show evolutionary divergence and identify AMP as a switch regulating glycolysis versus gluconeogenesis.FEBS J. 2020 Jul;287(13):2847-2861. doi: 10.1111/febs.15177. Epub 2020 Jan 8. FEBS J. 2020. PMID: 31838765 Free PMC article.
-
Structural and Functional Insights into Human Nuclear Cyclophilins.Biomolecules. 2018 Dec 4;8(4):161. doi: 10.3390/biom8040161. Biomolecules. 2018. PMID: 30518120 Free PMC article. Review.
References
-
- Zeidan E, Kepley CL, Sayes C and Sandros MG (2015) Surface plasmon resonance: a label‐free tool for cellular analysis. Nanomedicine 10, 1833–1846. - PubMed
-
- Lee TH, Hirst DJ and Aguilar MI (2015) New insights into the molecular mechanisms of biomembrane structural changes and interactions by optical biosensor technology. Biochim Biophys Acta 1848, 1868–1885. - PubMed
-
- Aristotelous T, Hopkins AL and Navratilova I (2015) Surface plasmon resonance analysis of seven‐transmembrane receptors. Methods Enzymol 556, 499–525. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

