SnoN Stabilizes the SMAD3/SMAD4 Protein Complex
- PMID: 28397834
- PMCID: PMC5387736
- DOI: 10.1038/srep46370
SnoN Stabilizes the SMAD3/SMAD4 Protein Complex
Abstract
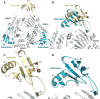
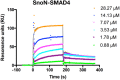
TGF-β signaling regulates cellular processes such as proliferation, differentiation and apoptosis through activation of SMAD transcription factors that are in turn modulated by members of the Ski-SnoN family. In this process, Ski has been shown to negatively modulate TGF-β signaling by disrupting active R-SMAD/Co-SMAD heteromers. Here, we show that the related regulator SnoN forms a stable complex with the R-SMAD (SMAD3) and the Co-SMAD (SMAD4). To rationalize this stabilization at the molecular level, we determined the crystal structure of a complex between the SAND domain of SnoN and the MH2-domain of SMAD4. This structure shows a binding mode that is compatible with simultaneous coordination of R-SMADs. Our results show that SnoN, and SMAD heteromers can form a joint structural core for the binding of other transcription modulators. The results are of fundamental importance for our understanding of the molecular mechanisms behind the modulation of TGF-β signaling.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Computational investigation of a ternary model of SnoN-SMAD3-SMAD4 complex.Comput Biol Chem. 2019 Dec;83:107159. doi: 10.1016/j.compbiolchem.2019.107159. Epub 2019 Nov 9. Comput Biol Chem. 2019. PMID: 31743832
-
Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation.Mol Cell Biol. 2007 Sep;27(17):6068-83. doi: 10.1128/MCB.00664-07. Epub 2007 Jun 25. Mol Cell Biol. 2007. PMID: 17591695 Free PMC article.
-
Two short segments of Smad3 are important for specific interaction of Smad3 with c-Ski and SnoN.J Biol Chem. 2003 Jan 3;278(1):531-6. doi: 10.1074/jbc.C200596200. Epub 2002 Nov 7. J Biol Chem. 2003. PMID: 12426322
-
[Role of Ski/SnoN protein in the regulation of TGF-beta signal pathway].Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003 Apr;25(2):233-6. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003. PMID: 12905729 Review. Chinese.
-
[Ski and SnoN: antagonistic proteins of TGFbeta signaling].Bull Cancer. 2000 Feb;87(2):135-7. Bull Cancer. 2000. PMID: 10705283 Review. French.
Cited by
-
Recent progress in metabolic reprogramming in gestational diabetes mellitus: a review.Front Endocrinol (Lausanne). 2024 Jan 3;14:1284160. doi: 10.3389/fendo.2023.1284160. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38234430 Free PMC article. Review.
-
Herbal compound 861 prevents hepatic fibrosis by inhibiting the TGF-β1/Smad/SnoN pathway in bile duct-ligated rats.BMC Complement Altern Med. 2018 Feb 5;18(1):52. doi: 10.1186/s12906-018-2119-7. BMC Complement Altern Med. 2018. PMID: 29402324 Free PMC article.
-
The SUMO System and TGFβ Signaling Interplay in Regulation of Epithelial-Mesenchymal Transition: Implications for Cancer Progression.Cancers (Basel). 2018 Aug 8;10(8):264. doi: 10.3390/cancers10080264. Cancers (Basel). 2018. PMID: 30096838 Free PMC article. Review.
-
Nucleotide substitutions at the p.Gly117 and p.Thr180 mutational hot-spots of SKI alter molecular dynamics and may affect cell cycle.J Hum Genet. 2024 Jan;69(1):53-58. doi: 10.1038/s10038-023-01193-7. Epub 2023 Sep 12. J Hum Genet. 2024. PMID: 37697026
-
Large-Scale Proteomics Data Reveal Integrated Prognosis-Related Protein Signatures and Role of SMAD4 and RAD50 in Prognosis and Immune Infiltrations of Prostate Cancer Microenvironment.Phenomics. 2022 Sep 27;2(6):404-418. doi: 10.1007/s43657-022-00070-1. eCollection 2022 Dec. Phenomics. 2022. PMID: 36939777 Free PMC article.
References
-
- Schmierer B. & Hill C. S. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8, 970–982 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

