Amyloid Fibrils from Hemoglobin
- PMID: 28398221
- PMCID: PMC5485726
- DOI: 10.3390/biom7020037
Amyloid Fibrils from Hemoglobin
Abstract
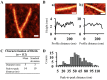
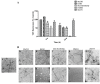
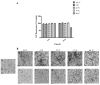
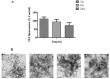
Amyloid fibrils are a class of insoluble protein nanofibers that are formed via the self-assembly of a wide range of peptides and proteins. They are increasingly exploited for a broad range of applications in bionanotechnology, such as biosensing and drug delivery, as nanowires, hydrogels, and thin films. Amyloid fibrils have been prepared from many proteins, but there has been no definitive characterization of amyloid fibrils from hemoglobin to date. Here, nanofiber formation was carried out under denaturing conditions using solutions of apo-hemoglobin extracted from bovine waste blood. A characteristic amyloid fibril morphology was confirmed by transmission electron microscopy (TEM) and atomic force microscopy (AFM), with mean fibril dimensions of approximately 5 nm diameter and up to several microns in length. The thioflavin T assay confirmed the presence of β-sheet structures in apo-hemoglobin fibrils, and X-ray fiber diffraction showed the characteristic amyloid cross-β quaternary structure. Apo-hemoglobin nanofibers demonstrated high stability over a range of temperatures (-20 to 80 °C) and pHs (2-10), and were stable in the presence of organic solvents and trypsin, confirming their potential as nanomaterials with versatile applications. This study conclusively demonstrates the formation of amyloid fibrils from hemoglobin for the first time, and also introduces a cost-effective method for amyloid fibril manufacture using meat industry by-products.
Keywords: amyloid fibrils; hemoglobin; nanofiber; nanofibril.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hughes V.A., Dunstan D.E. Amyloid fibrils - self-assembling proteins. Mod. Polym. Sci. 2009:559–594.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

