MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis
- PMID: 28398509
- PMCID: PMC5499743
- DOI: 10.1093/nar/gkx234
MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis
Abstract
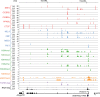
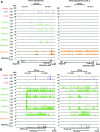
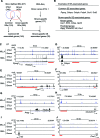
Histone H3K4me1/2 methyltransferases MLL3/MLL4 and H3K27 acetyltransferases CBP/p300 are major enhancer epigenomic writers. To understand how these epigenomic writers orchestrate enhancer landscapes in cell differentiation, we have profiled genomic binding of MLL4, CBP, lineage-determining transcription factors (EBF2, C/EBPβ, C/EBPα, PPARγ), coactivator MED1, RNA polymerase II, as well as epigenome (H3K4me1/2/3, H3K9me2, H3K27me3, H3K36me3, H3K27ac), transcriptome and chromatin opening during adipogenesis of immortalized preadipocytes derived from mouse brown adipose tissue (BAT). We show that MLL4 and CBP drive the dynamic enhancer epigenome, which correlates with the dynamic transcriptome. MLL3/MLL4 are required for CBP/p300 binding on enhancers activated during adipogenesis. Further, MLL4 and CBP identify super-enhancers (SEs) of adipogenesis and that MLL3/MLL4 are required for SE formation. Finally, in brown adipocytes differentiated in culture, MLL4 identifies primed SEs of genes fully activated in BAT such as Ucp1. Comparison of MLL4-defined SEs in brown and white adipogenesis identifies brown-specific SE-associated genes that could be involved in BAT functions. These results establish MLL3/MLL4 and CBP/p300 as master enhancer epigenomic writers and suggest that enhancer-priming by MLL3/MLL4 followed by enhancer-activation by CBP/p300 sequentially shape dynamic enhancer landscapes during cell differentiation. Our data also provide a rich resource for understanding epigenomic regulation of brown adipogenesis.
Published by Oxford University Press on behalf of Nucleic Acids Research 2017.
Figures






Similar articles
-
Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition.Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):11871-11876. doi: 10.1073/pnas.1606857113. Epub 2016 Oct 3. Proc Natl Acad Sci U S A. 2016. PMID: 27698142 Free PMC article.
-
A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription.Mol Cell. 2017 Jul 20;67(2):308-321.e6. doi: 10.1016/j.molcel.2017.06.028. Mol Cell. 2017. PMID: 28732206 Free PMC article.
-
Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis.Nat Commun. 2017 Dec 20;8(1):2217. doi: 10.1038/s41467-017-02403-5. Nat Commun. 2017. PMID: 29263365 Free PMC article.
-
Genomic occupancy of the transcriptional co-activators p300 and CBP.Transcription. 2013 Jan-Feb;4(1):18-23. doi: 10.4161/trns.22601. Epub 2012 Nov 6. Transcription. 2013. PMID: 23131664 Free PMC article. Review.
-
The MLL3/4 H3K4 methyltransferase complex in establishing an active enhancer landscape.Biochem Soc Trans. 2021 Jun 30;49(3):1041-1054. doi: 10.1042/BST20191164. Biochem Soc Trans. 2021. PMID: 34156443 Free PMC article. Review.
Cited by
-
Establishment and function of chromatin modification at enhancers.Open Biol. 2020 Oct;10(10):200255. doi: 10.1098/rsob.200255. Epub 2020 Oct 14. Open Biol. 2020. PMID: 33050790 Free PMC article.
-
Critical roles of transcriptional coactivator MED1 in the formation and function of mouse adipose tissues.Genes Dev. 2021 May 1;35(9-10):729-748. doi: 10.1101/gad.346791.120. Epub 2021 Apr 22. Genes Dev. 2021. PMID: 33888560 Free PMC article.
-
KMT2D-mediated H3K4me1 recruits YBX1 to facilitate triple-negative breast cancer progression through epigenetic activation of c-Myc.Clin Transl Med. 2024 Jul;14(7):e1753. doi: 10.1002/ctm2.1753. Clin Transl Med. 2024. PMID: 38967349 Free PMC article.
-
KMT2D deficiency drives lung squamous cell carcinoma and hypersensitivity to RTK-RAS inhibition.Cancer Cell. 2023 Jan 9;41(1):88-105.e8. doi: 10.1016/j.ccell.2022.11.015. Epub 2022 Dec 15. Cancer Cell. 2023. PMID: 36525973 Free PMC article.
-
Role of primary aging hallmarks in Alzheimer´s disease.Theranostics. 2023 Jan 1;13(1):197-230. doi: 10.7150/thno.79535. eCollection 2023. Theranostics. 2023. PMID: 36593969 Free PMC article. Review.
References
-
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. et al. . Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

