The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast
- PMID: 28398510
- PMCID: PMC5449591
- DOI: 10.1093/nar/gkx221
The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast
Abstract
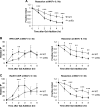
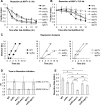
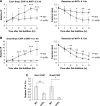
DNA double-strand breaks (DSBs) are repaired by either the non-homologous end joining (NHEJ) or homologous recombination (HR) pathway. Pathway choice is determined by the generation of 3΄ single-strand DNA overhangs at the break that are initiated by the action of the Mre11-Rad50-Xrs2 (MRX) complex to direct repair toward HR. DSB repair occurs in the context of chromatin, and multiple chromatin regulators have been shown to play important roles in the repair process. We have investigated the role of the SWI/SNF ATP-dependent nucleosome-remodeling complex in the repair of a defined DNA DSB. SWI/SNF was previously shown to regulate presynaptic events in HR, but its function in these events is unknown. We find that in the absence of functional SWI/SNF, the initiation of DNA end resection is significantly delayed. The delay in resection initiation is accompanied by impaired recruitment of MRX to the DSB, and other functions of MRX in HR including the recruitment of long-range resection factors and activation of the DNA damage response are also diminished. These phenotypes are correlated with a delay in the eviction of nucleosomes surrounding the DSB. We propose that SWI/SNF orchestrates the recruitment of a pool of MRX that is specifically dedicated to HR.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Processing of DNA double-stranded breaks and intermediates of recombination and repair by Saccharomyces cerevisiae Mre11 and its stimulation by Rad50, Xrs2, and Sae2 proteins.J Biol Chem. 2013 Apr 19;288(16):11273-86. doi: 10.1074/jbc.M112.439315. Epub 2013 Feb 26. J Biol Chem. 2013. PMID: 23443654 Free PMC article.
-
Xrs2 Dependent and Independent Functions of the Mre11-Rad50 Complex.Mol Cell. 2016 Oct 20;64(2):405-415. doi: 10.1016/j.molcel.2016.09.011. Epub 2016 Oct 13. Mol Cell. 2016. PMID: 27746018 Free PMC article.
-
The SWI/SNF ATPase BRG1 stimulates DNA end resection and homologous recombination by reducing nucleosome density at DNA double strand breaks and by promoting the recruitment of the CtIP nuclease.Cell Cycle. 2020 Nov;19(22):3096-3114. doi: 10.1080/15384101.2020.1831256. Epub 2020 Oct 12. Cell Cycle. 2020. PMID: 33044911 Free PMC article.
-
Structure-function relationships of the Mre11 protein in the control of DNA end bridging and processing.Curr Genet. 2019 Feb;65(1):11-16. doi: 10.1007/s00294-018-0861-5. Epub 2018 Jun 19. Curr Genet. 2019. PMID: 29922906 Review.
-
The role of the SWI/SNF chromatin remodelling complex in the response to DNA double strand breaks.DNA Repair (Amst). 2020 Sep;93:102919. doi: 10.1016/j.dnarep.2020.102919. DNA Repair (Amst). 2020. PMID: 33087260 Review.
Cited by
-
The COMPASS subunit Spp1 protects nascent DNA at the Tus/Ter replication fork barrier by limiting DNA availability to nucleases.Nat Commun. 2023 Sep 5;14(1):5430. doi: 10.1038/s41467-023-41100-4. Nat Commun. 2023. PMID: 37669924 Free PMC article.
-
Dpb4 promotes resection of DNA double-strand breaks and checkpoint activation by acting in two different protein complexes.Nat Commun. 2021 Aug 6;12(1):4750. doi: 10.1038/s41467-021-25090-9. Nat Commun. 2021. PMID: 34362907 Free PMC article.
-
The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts.Int J Mol Sci. 2017 Aug 16;18(8):1776. doi: 10.3390/ijms18081776. Int J Mol Sci. 2017. PMID: 28812991 Free PMC article. Review.
-
Multiple functions of SWI/SNF chromatin remodeling complex in plant-pathogen interactions.Stress Biol. 2021 Dec 9;1(1):18. doi: 10.1007/s44154-021-00019-w. Stress Biol. 2021. PMID: 37676626 Free PMC article. Review.
-
Sequence and chromatin features guide DNA double-strand break resection initiation.Mol Cell. 2023 Apr 20;83(8):1237-1250.e15. doi: 10.1016/j.molcel.2023.02.010. Epub 2023 Mar 13. Mol Cell. 2023. PMID: 36917982 Free PMC article.
References
-
- Mladenov E., Magin S., Soni A., Iliakis G.. Seminars in cancer biology. Semin. Cancer Biol. 2016; 37-38:51–64. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

