Inhibition of CRM1-mediated nuclear export of influenza A nucleoprotein and nuclear export protein as a novel target for antiviral drug development
- PMID: 28399435
- PMCID: PMC7111614
- DOI: 10.1016/j.virol.2017.04.001
Inhibition of CRM1-mediated nuclear export of influenza A nucleoprotein and nuclear export protein as a novel target for antiviral drug development
Abstract
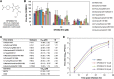
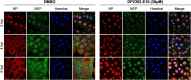
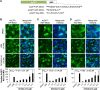
An anti-influenza compound, DP2392-E10 based on inhibition of the nuclear export function of the viral nucleoprotein-nuclear export signal 3 (NP-NES3) domain was successfully identified by our previous high-throughput screening system. Here, we demonstrated that DP2392-E10 exerts its antiviral effect by inhibiting replication of a broad range of influenza A subtypes. In regard to the molecular mechanism, we revealed that DP2392-E10 inhibits nuclear export of both viral NP and nuclear export protein (NEP). More specifically, in vitro pull-down assays revealed that DP2392-E10 directly binds cellular CRM1, which mediates nuclear export of NP and NEP. In silico docking suggested that DP2392-E10 binds at a region close to the HEAT9 and HEAT10 domains of CRM1. Together, these results indicate that the CRM1-mediated nuclear export function of influenza virus represents a new potential target for antiviral drug development, and also provide a core structure for a novel class of inhibitors that target this function.
Keywords: Chromosome region maintenance 1; Influenza A virus; Nuclear export protein; Nuclear export signal; Nucleoprotein.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Naproxen Exhibits Broad Anti-influenza Virus Activity in Mice by Impeding Viral Nucleoprotein Nuclear Export.Cell Rep. 2019 May 7;27(6):1875-1885.e5. doi: 10.1016/j.celrep.2019.04.053. Cell Rep. 2019. PMID: 31067470
-
NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway.Viruses. 2016 Jul 28;8(8):209. doi: 10.3390/v8080209. Viruses. 2016. PMID: 27483302 Free PMC article.
-
Inhibition of RAN attenuates influenza a virus replication and nucleoprotein nuclear export.Emerg Microbes Infect. 2024 Dec;13(1):2387910. doi: 10.1080/22221751.2024.2387910. Epub 2024 Aug 12. Emerg Microbes Infect. 2024. PMID: 39087696 Free PMC article.
-
Antiviral drug development by targeting RNA binding site, oligomerization and nuclear export of influenza nucleoprotein.Int J Biol Macromol. 2024 Dec;282(Pt 4):136996. doi: 10.1016/j.ijbiomac.2024.136996. Epub 2024 Oct 31. Int J Biol Macromol. 2024. PMID: 39486729 Review.
-
Nuclear Export as a Novel Therapeutic Target: The CRM1 Connection.Curr Cancer Drug Targets. 2015;15(7):575-92. doi: 10.2174/156800961507150828223554. Curr Cancer Drug Targets. 2015. PMID: 26324128 Review.
Cited by
-
Phosphorylation Status of Tyrosine 78 Residue Regulates the Nuclear Export and Ubiquitination of Influenza A Virus Nucleoprotein.Front Microbiol. 2019 Aug 7;10:1816. doi: 10.3389/fmicb.2019.01816. eCollection 2019. Front Microbiol. 2019. PMID: 31440228 Free PMC article.
-
Anti-Influenza Drug Discovery and Development: Targeting the Virus and Its Host by All Possible Means.Adv Exp Med Biol. 2021;1322:195-218. doi: 10.1007/978-981-16-0267-2_8. Adv Exp Med Biol. 2021. PMID: 34258742
-
Loxapine inhibits replication of hepatitis A virus in vitro and in vivo by targeting viral protein 2C.PLoS Pathog. 2024 Mar 13;20(3):e1012091. doi: 10.1371/journal.ppat.1012091. eCollection 2024 Mar. PLoS Pathog. 2024. PMID: 38478584 Free PMC article.
-
Unique Mode of Antiviral Action of a Marine Alkaloid against Ebola Virus and SARS-CoV-2.Viruses. 2022 Apr 15;14(4):816. doi: 10.3390/v14040816. Viruses. 2022. PMID: 35458549 Free PMC article.
-
Host-Directed Antiviral Therapy.Clin Microbiol Rev. 2020 May 13;33(3):e00168-19. doi: 10.1128/CMR.00168-19. Print 2020 Jun 17. Clin Microbiol Rev. 2020. PMID: 32404434 Free PMC article. Review.
References
-
- Aida Y., Sasaki Y., Hagiwara K. Discovery of novel antiviral agents directed against the influenza A Virus Nucleoprotein. In: Arbuthnot P., editor. Antiviral Drugs - Aspects of Clinical Use and Recent Advances. InTech; Rijeka, Croatia: 2012. pp. 99–120.
-
- Cai W., Li Y., Chen S., Wang M., Zhang A., Zhou H., Chen H., Jin M. 14-Deoxy-11,12-dehydroandrographolide exerts anti-influenza A virus activity and inhibits replication of H5N1 virus by restraining nuclear export of viral ribonucleoprotein complexes. Antivir. Res. 2015;118:82–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

