A machine learning approach for viral genome classification
- PMID: 28399797
- PMCID: PMC5387389
- DOI: 10.1186/s12859-017-1602-3
A machine learning approach for viral genome classification
Abstract
Background: Advances in cloning and sequencing technology are yielding a massive number of viral genomes. The classification and annotation of these genomes constitute important assets in the discovery of genomic variability, taxonomic characteristics and disease mechanisms. Existing classification methods are often designed for specific well-studied family of viruses. Thus, the viral comparative genomic studies could benefit from more generic, fast and accurate tools for classifying and typing newly sequenced strains of diverse virus families.
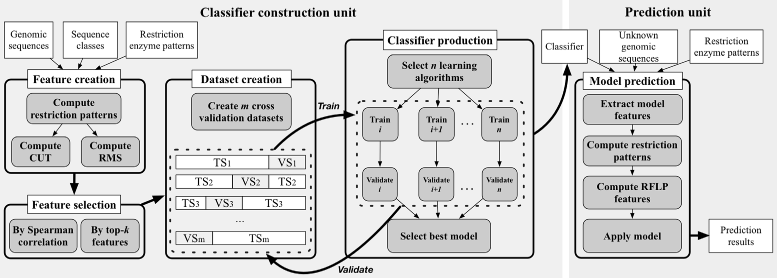
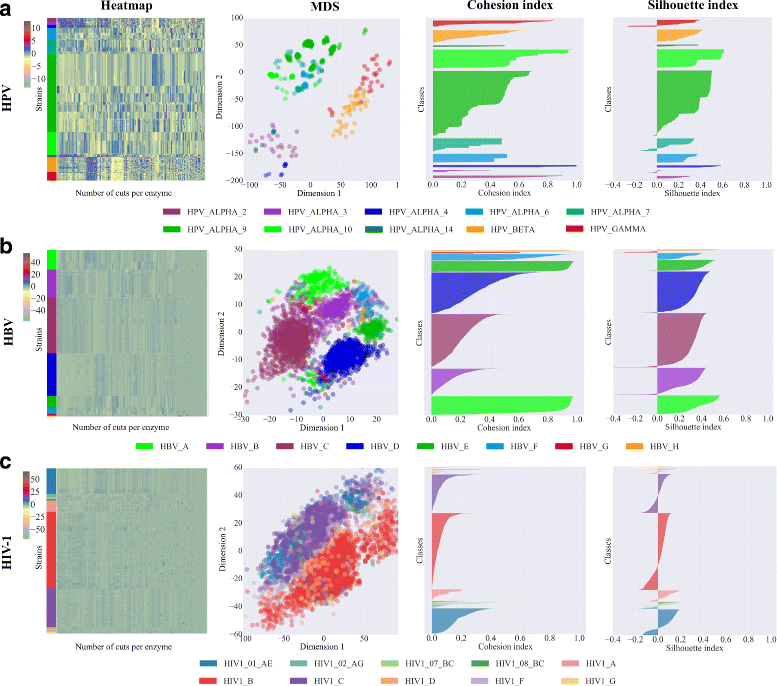
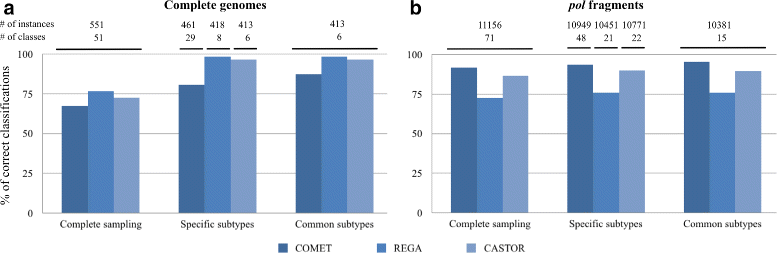
Results: Here, we introduce a virus classification platform, CASTOR, based on machine learning methods. CASTOR is inspired by a well-known technique in molecular biology: restriction fragment length polymorphism (RFLP). It simulates, in silico, the restriction digestion of genomic material by different enzymes into fragments. It uses two metrics to construct feature vectors for machine learning algorithms in the classification step. We benchmark CASTOR for the classification of distinct datasets of human papillomaviruses (HPV), hepatitis B viruses (HBV) and human immunodeficiency viruses type 1 (HIV-1). Results reveal true positive rates of 99%, 99% and 98% for HPV Alpha species, HBV genotyping and HIV-1 M subtyping, respectively. Furthermore, CASTOR shows a competitive performance compared to well-known HIV-1 specific classifiers (REGA and COMET) on whole genomes and pol fragments.
Conclusion: The performance of CASTOR, its genericity and robustness could permit to perform novel and accurate large scale virus studies. The CASTOR web platform provides an open access, collaborative and reproducible machine learning classifiers. CASTOR can be accessed at http://castor.bioinfo.uqam.ca .
Keywords: Prediction; Sequence classification; Virus classification.
Figures

Similar articles
-
ML-DSP: Machine Learning with Digital Signal Processing for ultrafast, accurate, and scalable genome classification at all taxonomic levels.BMC Genomics. 2019 Apr 3;20(1):267. doi: 10.1186/s12864-019-5571-y. BMC Genomics. 2019. PMID: 30943897 Free PMC article.
-
Toward an Alignment-Free Method for Feature Extraction and Accurate Classification of Viral Sequences.J Comput Biol. 2019 Jun;26(6):519-535. doi: 10.1089/cmb.2018.0239. Epub 2019 May 3. J Comput Biol. 2019. PMID: 31050550
-
An open-source k-mer based machine learning tool for fast and accurate subtyping of HIV-1 genomes.PLoS One. 2018 Nov 14;13(11):e0206409. doi: 10.1371/journal.pone.0206409. eCollection 2018. PLoS One. 2018. PMID: 30427878 Free PMC article.
-
Unveiling the ghost: machine learning's impact on the landscape of virology.J Gen Virol. 2025 Jan;106(1). doi: 10.1099/jgv.0.002067. J Gen Virol. 2025. PMID: 39804261 Review.
-
Machine learning and its applications in plant molecular studies.Brief Funct Genomics. 2020 Jan 22;19(1):40-48. doi: 10.1093/bfgp/elz036. Brief Funct Genomics. 2020. PMID: 31867668 Review.
Cited by
-
Convolutional Neural Network Applied to SARS-CoV-2 Sequence Classification.Sensors (Basel). 2022 Jul 31;22(15):5730. doi: 10.3390/s22155730. Sensors (Basel). 2022. PMID: 35957287 Free PMC article.
-
Beyond cells - The virome in the human holobiont.Microb Cell. 2019 Jul 1;6(9):373-396. doi: 10.15698/mic2019.09.689. Microb Cell. 2019. PMID: 31528630 Free PMC article. Review.
-
WalkIm: Compact image-based encoding for high-performance classification of biological sequences using simple tuning-free CNNs.PLoS One. 2022 Apr 15;17(4):e0267106. doi: 10.1371/journal.pone.0267106. eCollection 2022. PLoS One. 2022. PMID: 35427371 Free PMC article.
-
Heuristic Analysis of Genomic Sequence Processing Models for High Efficiency Prediction: A Statistical Perspective.Curr Genomics. 2022 Nov 18;23(5):299-317. doi: 10.2174/1389202923666220927105311. Curr Genomics. 2022. PMID: 36778194 Free PMC article. Review.
-
Automated classification of giant virus genomes using a random forest model built on trademark protein families.Npj Viruses. 2024 Mar 8;2(1):9. doi: 10.1038/s44298-024-00021-9. Npj Viruses. 2024. PMID: 40295679 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

