Next-generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia
- PMID: 28399885
- PMCID: PMC5387353
- DOI: 10.1186/s13045-017-0450-y
Next-generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia
Abstract
Background: Chronic lymphocytic leukemia (CLL) is a highly genetically heterogeneous disease. Although CLL has been traditionally considered as a mature B cell leukemia, few independent studies have shown that the genetic alterations may appear in CD34+ hematopoietic progenitors. However, the presence of both chromosomal aberrations and gene mutations in CD34+ cells from the same patients has not been explored.
Methods: Amplicon-based deep next-generation sequencing (NGS) studies were carried out in magnetically activated-cell-sorting separated CD19+ mature B lymphocytes and CD34+ hematopoietic progenitors (n = 56) to study the mutational status of TP53, NOTCH1, SF3B1, FBXW7, MYD88, and XPO1 genes. In addition, ultra-deep NGS was performed in a subset of seven patients to determine the presence of mutations in flow-sorted CD34+CD19- early hematopoietic progenitors. Fluorescence in situ hybridization (FISH) studies were performed in the CD34+ cells from nine patients of the cohort to examine the presence of cytogenetic abnormalities.
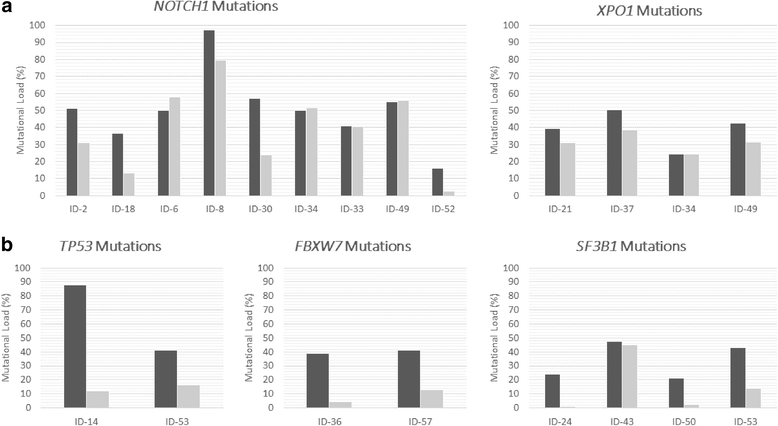
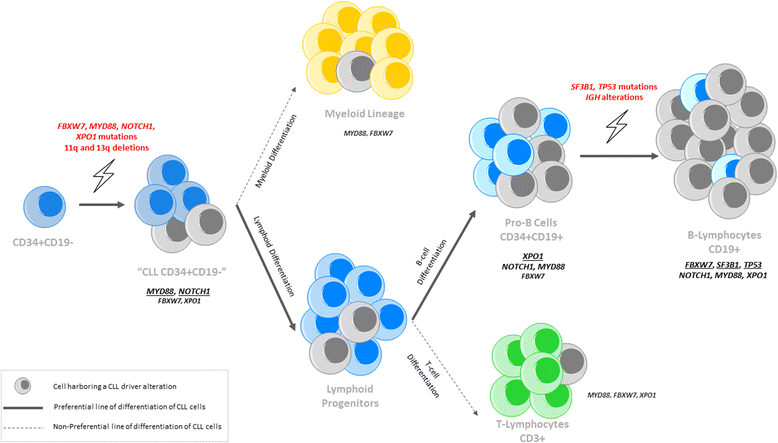
Results: NGS studies revealed a total of 28 mutations in 24 CLL patients. Interestingly, 15 of them also showed the same mutations in their corresponding whole population of CD34+ progenitors. The majority of NOTCH1 (7/9) and XPO1 (4/4) mutations presented a similar mutational burden in both cell fractions; by contrast, mutations of TP53 (2/2), FBXW7 (2/2), and SF3B1 (3/4) showed lower mutational allele frequencies, or even none, in the CD34+ cells compared with the CD19+ population. Ultra-deep NGS confirmed the presence of FBXW7, MYD88, NOTCH1, and XPO1 mutations in the subpopulation of CD34+CD19- early hematopoietic progenitors (6/7). Furthermore, FISH studies showed the presence of 11q and 13q deletions (2/2 and 3/5, respectively) in CD34+ progenitors but the absence of IGH cytogenetic alterations (0/2) in the CD34+ cells. Combining all the results from NGS and FISH, a model of the appearance and expansion of genetic alterations in CLL was derived, suggesting that most of the genetic events appear on the hematopoietic progenitors, although these mutations could induce the beginning of tumoral cell expansion at different stage of B cell differentiation.
Conclusions: Our study showed the presence of both gene mutations and chromosomal abnormalities in early hematopoietic progenitor cells from CLL patients.
Keywords: Chromosomal abnormality; Chronic lymphocytic leukemia; FISH; Hematopoietic progenitors; Mutation; Next-generation sequencing.
Figures

References
-
- Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–56. doi: 10.1182/blood-2007-06-093906. - DOI - PMC - PubMed
-
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

