TLR sensing of bacterial spore-associated RNA triggers host immune responses with detrimental effects
- PMID: 28400473
- PMCID: PMC5413331
- DOI: 10.1084/jem.20161141
TLR sensing of bacterial spore-associated RNA triggers host immune responses with detrimental effects
Abstract
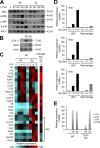
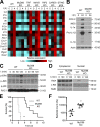
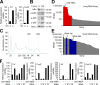
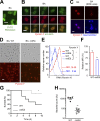
The spores of pathogenic bacteria are involved in host entry and the initial encounter with the host immune system. How bacterial spores interact with host immunity, however, remains poorly understood. Here, we show that the spores of Bacillus anthracis (BA), the etiologic agent of anthrax, possess an intrinsic ability to induce host immune responses. This immunostimulatory activity is attributable to high amounts of RNA present in the spore surface layer. RNA-sensing TLRs, TLR7, and TLR13 in mice and their human counterparts, are responsible for detecting and triggering the host cell response to BA spores, whereas TLR2 mediates the sensing of vegetative BA. BA spores, but not vegetative BA, induce type I IFN (IFN-I) production. Although TLR signaling in itself affords protection against BA, spore RNA-induced IFN-I signaling is disruptive to BA clearance. Our study suggests a role for bacterial spore-associated RNA in microbial pathogenesis and illustrates a little known aspect of interactions between the host and spore-forming bacteria.
© 2017 Choo et al.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

