Streptococcus agalactiae Causing Neonatal Infections in Portugal (2005-2015): Diversification and Emergence of a CC17/PI-2b Multidrug Resistant Sublineage
- PMID: 28400757
- PMCID: PMC5368217
- DOI: 10.3389/fmicb.2017.00499
Streptococcus agalactiae Causing Neonatal Infections in Portugal (2005-2015): Diversification and Emergence of a CC17/PI-2b Multidrug Resistant Sublineage
Abstract
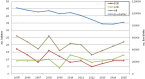
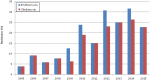
The molecular characterization of 218 GBS isolates recovered from neonatal invasive infections in Portugal in 2005-2015 revealed the existence of a small number of genetically distinct lineages that were present over a significant time-span. Serotypes III and Ia were dominant in the population, together accounting for >80% of the isolates. Clonal complex 17 included 50% of all isolates, highlighting the importance of the hypervirulent genetic lineage represented by serotype III ST17/rib/PI-1+PI-2b. Serotype Ia was represented mainly by ST23, previously reported as dominant among invasive disease in non-pregnant adults in Portugal, but also by ST24, showing an increased frequency among late-onset disease. Overall erythromycin resistance was 16%, increasing during the study period (p < 0.001). Macrolide resistance was overrepresented among CC1 and CC19 isolates (p < 0.001 and p = 0.008, respectively). While representatives of the hypervirulent CC17 lineage were mostly susceptible to macrolides, we identified for the first time in Europe a recently emerging sublineage characterized by the loss of PI-1 (CC17/PI-2b), simultaneously resistant to macrolides, lincosamides, and tetracycline, also exhibiting high-level resistance to streptomycin and kanamycin. The stability and dominance of CC17 among neonatal invasive infections in the past decades indicates that it is extremely well adapted to its niche; however emerging resistance in this genetic background may have significant implications for the prevention and management of GBS disease.
Keywords: CC17; Streptococcus agalactiae; antimicrobial resistance; invasive disease; neonates.
Figures
References
-
- Bekker V., Bijlsma M. W., van de Beek D., Kuijpers T. W., van der Ende A. (2014). Incidence of invasive group B streptococcal disease and pathogen genotype distribution in newborn babies in the Netherlands over 25 years: a nationwide surveillance study. Lancet Infect. Dis. 14, 1083–1089. 10.1016/S1473-3099(14)70919-3 - DOI - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approch to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300.
-
- Campisi E., Rosini R., Ji W., Guidotti S., Rojas-López M., Geng G., et al. . (2016). Genomic analysis reveals multi-drug resistance clusters in Group B Streptococcus CC17 hypervirulent isolates causing neonatal invasive disease in Southern Mainland China. Front. Microbiol. 7:1265. 10.3389/fmicb.2016.01265 - DOI - PMC - PubMed
-
- Carrico J. A., Silva-Costa C., Melo-Cristino J., Pinto F. R., de Lencastre H., Almeida J. S., et al. . (2006). Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44, 2524–2532. 10.1128/JCM.02536-05 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous