Regulation of miR163 and its targets in defense against Pseudomonas syringae in Arabidopsis thaliana
- PMID: 28401908
- PMCID: PMC5388894
- DOI: 10.1038/srep46433
Regulation of miR163 and its targets in defense against Pseudomonas syringae in Arabidopsis thaliana
Abstract
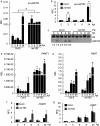
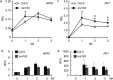
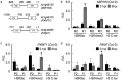
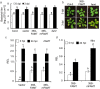
Small RNAs are important regulators for a variety of biological processes, including leaf development, flowering-time, embryogenesis and defense responses. miR163 is a non-conserved miRNA and its locus has evolved recently through inverted duplication of its target genes to which they belong to the SABATH family of related small-molecule methyltransferases (MTs). In Arabidopsis thaliana, previous study demonstrated that miR163 accumulation was induced by alamethicin treatment, suggesting its roles in defense response pathways. Enhanced resistance against Pseudomonas syringae pv. tomato (Pst) was observed in the mir163 mutant, whereas transgenic lines overexpressing miR163 showed increase sensitivity to Pst, suggesting that miR163 is a negative regulator of defense response. Elevated level of miR163 and its targets in A. thaliana were observed upon Pst treatment, suggesting a modulating relationship between miR163 and its targets. In addition, miR163 and histone deacetylase were found to act cooperatively in mediating defense against Pst. Transgenic plants overexpressing miR163-resistant targets suggested their different contributions in defense. Results from this study revealed that the stress-inducible miR163 and its targets act in concert to modulate defense responses against bacterial pathogen in A. thaliana.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

