BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes
- PMID: 28401936
- PMCID: PMC5638664
- DOI: 10.1038/mi.2017.24
BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes
Abstract
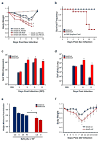
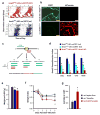
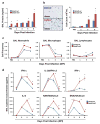
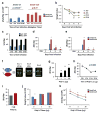
Patients with asthma experience circadian variations in their symptoms. However it remains unclear how specific aspects of this common airway disease relate to clock genes, which are critical to the generation of circadian rhythms in mammals. Here, we used a viral model of acute and chronic airway disease to examine how circadian clock disruption affects asthmatic lung phenotypes. Deletion of the core clock gene bmal1 or environmental disruption of circadian function by jet lag exacerbated acute viral bronchiolitis caused by Sendai virus (SeV) and influenza A virus in mice. Post-natal deletion of bmal1 was sufficient to trigger increased SeV susceptibility and correlated with impaired control of viral replication. Importantly, bmal1-/- mice developed much more extensive asthma-like airway changes post infection, including mucus production and increased airway resistance. In human airway samples from two asthma cohorts, we observed altered expression patterns of multiple clock genes. Our results suggest a role for bmal1 in the development of asthmatic airway disease via the regulation of lung antiviral responses to common viral triggers of asthma.
Conflict of interest statement
Figures



References
-
- Bergeron C, Al-Ramli W, Hamid Q. Remodeling in asthma. Proceedings of the American Thoracic Society. 2009;6(3):301–305. - PubMed
-
- Meissner HC. Viral Bronchiolitis in Children. The New England journal of medicine. 2016;374(18):1793–1794. - PubMed
-
- Corne JM, Marshall C, Smith S, Schreiber J, Sanderson G, Holgate ST, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359(9309):831–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL091762/HL/NHLBI NIH HHS/United States
- U10 HL109257/HL/NHLBI NIH HHS/United States
- UH2 HL123429/HL/NHLBI NIH HHS/United States
- R01 HL121791/HL/NHLBI NIH HHS/United States
- S10 RR027552/RR/NCRR NIH HHS/United States
- R01 HL069149/HL/NHLBI NIH HHS/United States
- R01 HL092486/HL/NHLBI NIH HHS/United States
- R01 HL135846/HL/NHLBI NIH HHS/United States
- T32 AI007172/AI/NIAID NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- R01 HL061895/HL/NHLBI NIH HHS/United States
- R01 AI130591/AI/NIAID NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- K08 GM102694/GM/NIGMS NIH HHS/United States
- UH3 HL123429/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

