Engineering and expressing circular RNAs via tRNA splicing
- PMID: 28402213
- PMCID: PMC5680671
- DOI: 10.1080/15476286.2017.1317911
Engineering and expressing circular RNAs via tRNA splicing
Abstract
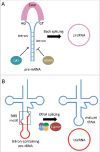
Circular (circ)RNAs have recently become a subject of great biologic interest. It is now clear that they represent a diverse and abundant class of RNAs with regulated expression and evolutionarily conserved functions. There are several mechanisms by which RNA circularization can occur in vivo. Here, we focus on the biogenesis of tRNA intronic circular RNAs (tricRNAs) in archaea and animals, and we detail their use as research tools for orthogonal, directed circRNA expression in vivo.
Keywords: Back-splicing; RNA aptamers; circRNA; ectopic gene expression; tRNA processing; tricRNA.
Figures
Similar articles
-
A Method for Expressing and Imaging Abundant, Stable, Circular RNAs In Vivo Using tRNA Splicing.Methods Enzymol. 2016;572:215-36. doi: 10.1016/bs.mie.2016.02.018. Epub 2016 Mar 25. Methods Enzymol. 2016. PMID: 27241756
-
Early history of circular RNAs, children of splicing.RNA Biol. 2017 Aug 3;14(8):975-977. doi: 10.1080/15476286.2016.1227903. Epub 2016 Aug 26. RNA Biol. 2017. PMID: 27563746 Free PMC article.
-
Circular RNAs: Unexpected outputs of many protein-coding genes.RNA Biol. 2017 Aug 3;14(8):1007-1017. doi: 10.1080/15476286.2016.1227905. Epub 2016 Aug 29. RNA Biol. 2017. PMID: 27571848 Free PMC article. Review.
-
Molecular determinants of metazoan tricRNA biogenesis.Nucleic Acids Res. 2019 Jul 9;47(12):6452-6465. doi: 10.1093/nar/gkz311. Nucleic Acids Res. 2019. PMID: 31032518 Free PMC article.
-
The Biogenesis, Functions, and Challenges of Circular RNAs.Mol Cell. 2018 Aug 2;71(3):428-442. doi: 10.1016/j.molcel.2018.06.034. Epub 2018 Jul 26. Mol Cell. 2018. PMID: 30057200 Review.
Cited by
-
Role of Virally Encoded Circular RNAs in the Pathogenicity of Human Oncogenic Viruses.Front Microbiol. 2021 Apr 20;12:657036. doi: 10.3389/fmicb.2021.657036. eCollection 2021. Front Microbiol. 2021. PMID: 33959113 Free PMC article. Review.
-
Circular RNAs and cervical cancer: friends or foes? A landscape on circRNA-mediated regulation of key signaling pathways involved in the onset and progression of HPV-related cervical neoplasms.Cell Commun Signal. 2024 Feb 10;22(1):107. doi: 10.1186/s12964-024-01494-0. Cell Commun Signal. 2024. PMID: 38341592 Free PMC article. Review.
-
Modified Nucleotides for Chemical and Enzymatic Synthesis of Therapeutic RNA.Curr Med Chem. 2023;30(11):1320-1347. doi: 10.2174/0929867330666221014111403. Curr Med Chem. 2023. PMID: 36239720
-
circRNA-miRNA-mRNA regulatory network in human lung cancer: an update.Cancer Cell Int. 2020 May 19;20:173. doi: 10.1186/s12935-020-01245-4. eCollection 2020. Cancer Cell Int. 2020. PMID: 32467668 Free PMC article. Review.
-
CircRNAs as promising biomarker in diagnosis of breast cancer: An updated meta-analysis.J Clin Lab Anal. 2021 Sep;35(9):e23934. doi: 10.1002/jcla.23934. Epub 2021 Jul 31. J Clin Lab Anal. 2021. PMID: 34331339 Free PMC article.
References
-
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/10.1038/nbt.2890 - DOI - PMC - PubMed
-
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; https://doi.org/10.1261/rna.035667.112 - DOI - PMC - PubMed
-
- Coqcuerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559; https://doi.org/10.1261/rna.047126.114 - DOI - PubMed
-
- Lasda E, Parker R. Circular RNAs: Diversity of form and function. RNA 2014; 20:1829-42; PMID:25404635; https://doi.org/10.1261/rna.047126.114 - DOI - PMC - PubMed
-
- Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 2015; 43:2454-65; PMID:25662225; https://doi.org/10.1093/nar/gkv045 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
