Amplification of a transgene within a long array of replication origins favors higher gene expression in animal cells
- PMID: 28403180
- PMCID: PMC5389822
- DOI: 10.1371/journal.pone.0175585
Amplification of a transgene within a long array of replication origins favors higher gene expression in animal cells
Abstract
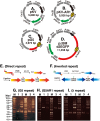
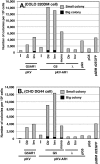
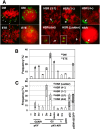
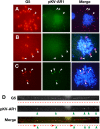
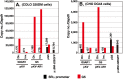
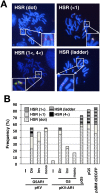
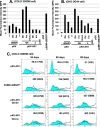
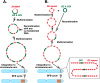
Plasmids with both a mammalian replication initiation region (IR) and a matrix attachment region (MAR) are spontaneously amplified in transfected cells, and generate extrachromosomal double minute (DM) or chromosomal homogeneously staining region (HSR). We previously isolated the shortest core IR (G5) required for gene amplification. In this study, we ligated the G5 DNA to create direct or inverted repeats, mixed the repeats with an expression plasmid, and transfected the mixture into human COLO 320DM or hamster CHO DG44 cells. Consequently, we found that the transfected sequence generated DMs or HSR where, surprisingly, the plasmid sequence was embedded within a long stretch of G5 sequences. The amplified structure from the direct G5 repeats was stable, whereas that from the inverted repeats was not. The amplification might be explained by the efficient replication/multimerization of the G5 repeat and recombination with the co-transfected plasmid in an extrachromosomal context. The product might then be integrated into a chromosome arm to generate a HSR. The expression from the plasmid within the long G5 array was much higher than that from a simple plasmid repeat. Because G5 is a core IR that favors gene expression, a long array of G5 provides an excellent environment for gene expression from the embedded plasmid.
Conflict of interest statement
Figures
References
-
- Omasa T. Gene Amplification and Its Application in Cell and Tissue Engineering. J Biosci Bioeng. 2002;94(6):600–605. - PubMed
-
- Shimizu N, Miura Y, Sakamoto Y, Tsutsui K. Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res. 2001;61(19):6987–6990. - PubMed
-
- Shimizu N, Hashizume T, Shingaki K, Kawamoto JK. Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer Res. 2003;63(17):5281–5290. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

