sRNAs as possible regulators of retrotransposon activity in Cryptococcus gattii VGII
- PMID: 28403818
- PMCID: PMC5389150
- DOI: 10.1186/s12864-017-3688-4
sRNAs as possible regulators of retrotransposon activity in Cryptococcus gattii VGII
Abstract
Background: The absence of Argonaute genes in the fungal pathogen Cryptococcus gattii R265 and other VGII strains indicates that yeasts of this genotype cannot have a functional RNAi pathway, an evolutionarily conserved gene silencing mechanism performed by small RNAs. The success of the R265 strain as a pathogen that caused the Pacific Northwest and Vancouver Island outbreaks may imply that RNAi machinery loss could be beneficial under certain circumstances during evolution. As a result, a hypermutant phenotype would be created with high rates of genome retrotransposition, for instance. This study therefore aimed to evaluate in silicio the effect of retrotransposons and their control mechanisms by small RNAs on genomic stability and synteny loss of C. gattii R265 through retrotransposons sequence comparison and orthology analysis with other 16 C. gattii genomic sequences available.
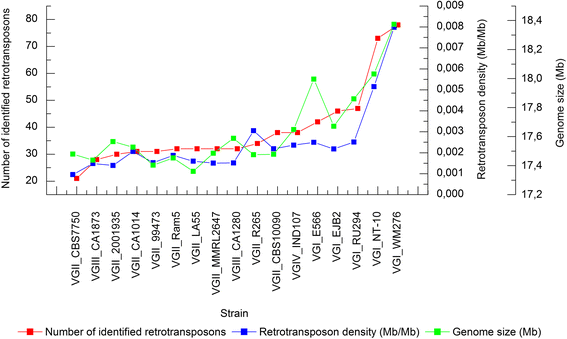
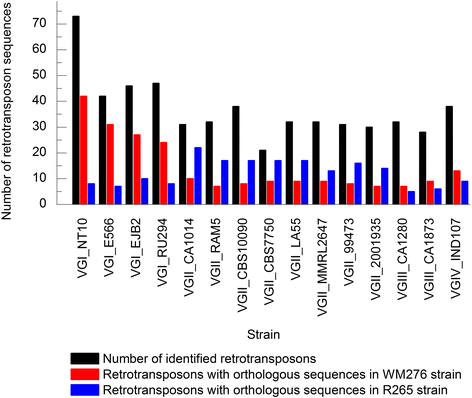
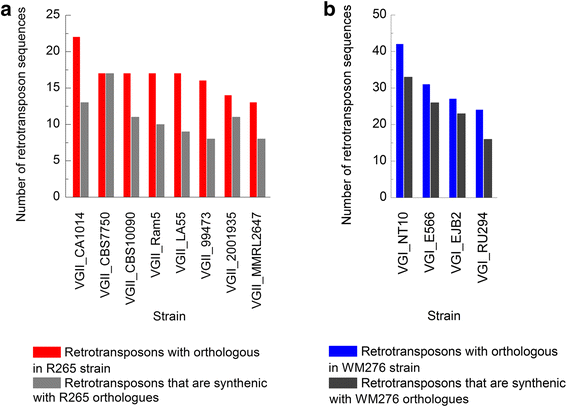
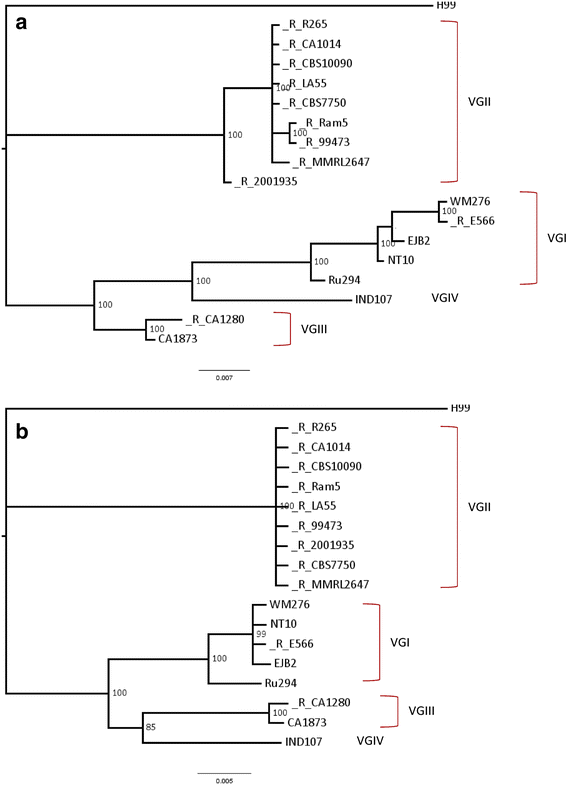
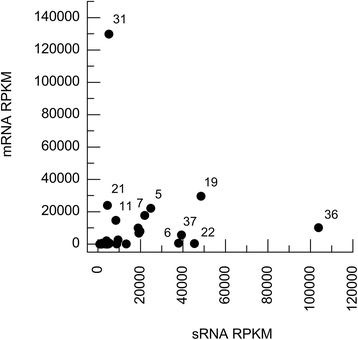
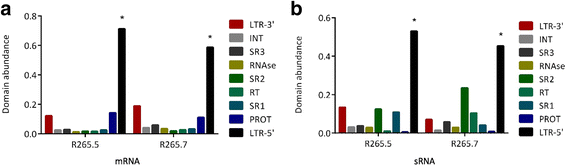
Results: Retrotransposon mining identified a higher sequence count to VGI genotype compared to VGII, VGIII, and VGIV. However, despite the lower retrotransposon number, VGII exhibited increased synteny loss and genome rearrangement events. RNA-Seq analysis indicated highly expressed retrotransposons as well as sRNA production.
Conclusions: Genome rearrangement and synteny loss may suggest a greater retrotransposon mobilization caused by RNAi pathway absence, but the effective presence of sRNAs that matches retrotransposon sequences means that an alternative retrotransposon silencing mechanism could be active in genomic integrity maintenance of C. gattii VGII strains.
Keywords: Cryptococcus gattii; RNAi; Retrotransposons; Synteny.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

