Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species
- PMID: 28403834
- PMCID: PMC5388996
- DOI: 10.1186/s12864-017-3682-x
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species
Abstract
Background: Nucleotide binding site (NBS) genes encode a large family of disease resistance (R) proteins in plants. The availability of genomic data of the two diploid cotton species, Gossypium arboreum and Gossypium raimondii, and the two allotetraploid cotton species, Gossypium hirsutum (TM-1) and Gossypium barbadense allow for a more comprehensive and systematic comparative study of NBS-encoding genes to elucidate the mechanisms of cotton disease resistance.
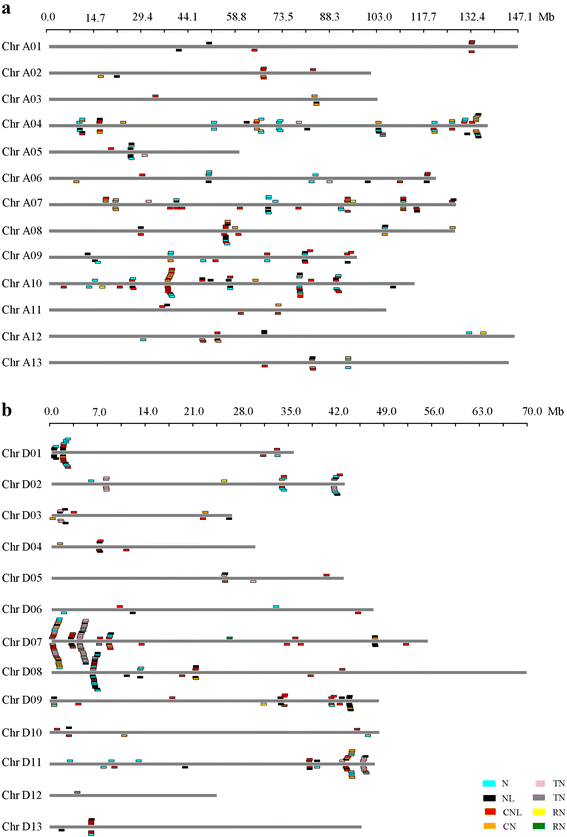
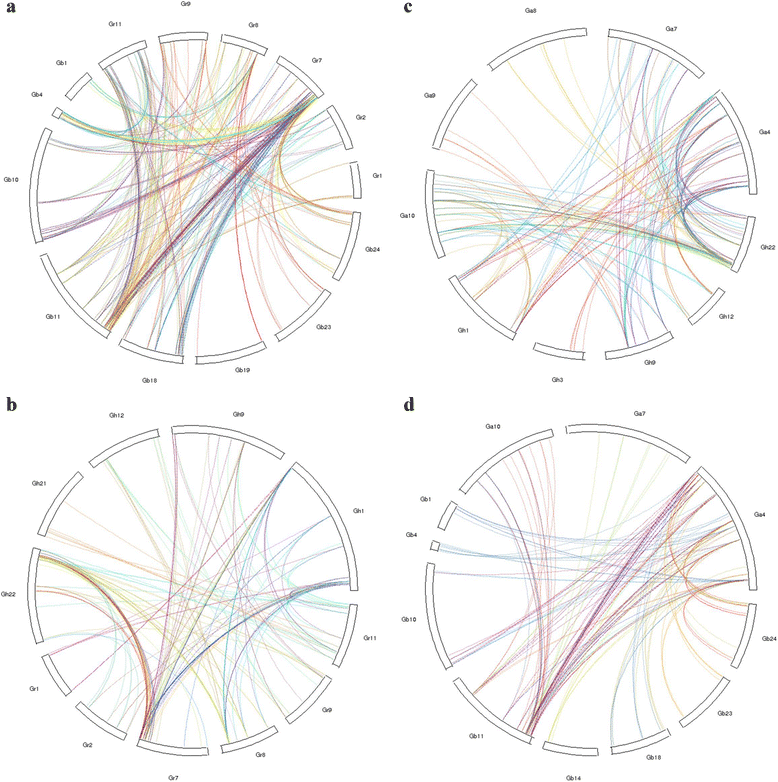
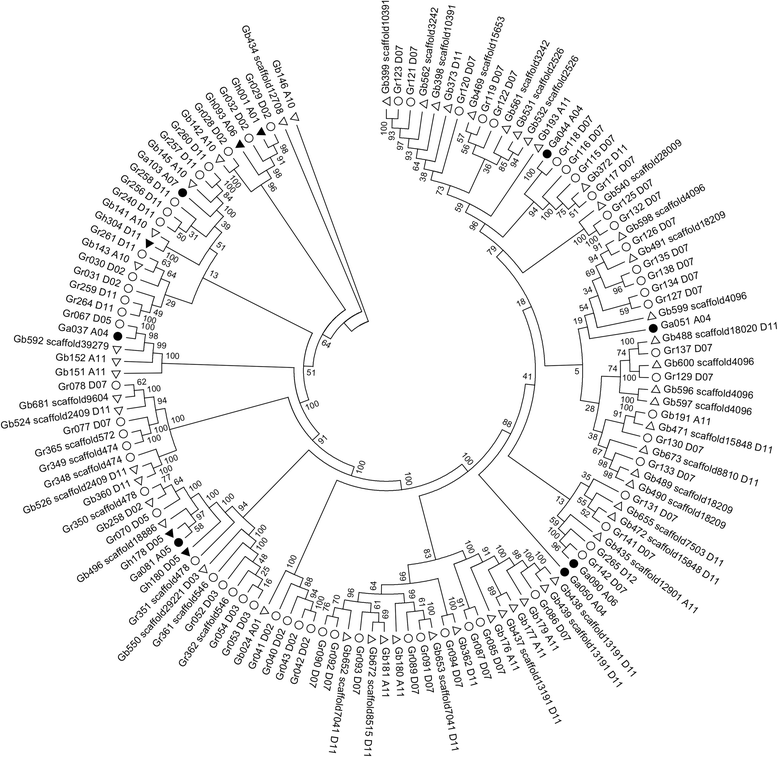
Results: Based on the genome assembly data, 246, 365, 588 and 682 NBS-encoding genes were identified in G. arboreum, G. raimondii, G. hirsutum and G. barbadense, respectively. The distribution of NBS-encoding genes among the chromosomes was nonrandom and uneven, and was tended to form clusters. Gene structure analysis showed that G. arboreum and G. hirsutum possessed a greater proportion of CN, CNL, and N genes and a lower proportion of NL, TN and TNL genes compared to that of G. raimondii and G. barbadense, while the percentages of RN and RNL genes remained relatively unchanged. The percentage changes among them were largest for TNL genes, about 7 times. Exon statistics showed that the average exon numbers per NBS gene in G. raimondii and G. barbadense were all greater than that in G. arboretum and G. hirsutum. Phylogenetic analysis revealed that the TIR-NBS genes of G. barbadense were closely related with that of G. raimondii. Sequence similarity analysis showed that diploid cotton G. arboreum possessed a larger proportion of NBS-encoding genes similar to that of allotetraploid cotton G. hirsutum, while diploid G. raimondii possessed a larger proportion of NBS-encoding genes similar to that of allotetraploid cotton G. barbadense. The synteny analysis showed that more NBS genes in G. raimondii and G. arboreum were syntenic with that in G. barbadense and G. hirsutum, respectively.
Conclusions: The structural architectures, amino acid sequence similarities and synteny of NBS-encoding genes between G. arboreum and G. hirsutum, and between G. raimondii and G. barbadense were the highest among comparisons between the diploid and allotetraploid genomes, indicating that G. hirsutum inherited more NBS-encoding genes from G. arboreum, while G. barbadense inherited more NBS-encoding genes from G. raimondii. This asymmetric evolution of NBS-encoding genes may help to explain why G. raimondii and G. barbadense are more resistant to Verticillium wilt, whereas G. arboreum and G. hirsutum are more susceptible to Verticillium wilt. The disease resistances of the allotetraploid cotton were related to their NBS-encoding genes especially in regard from which diploid progenitor they were derived, and the TNL genes may have a significant role in disease resistance to Verticillium wilt in G. raimondii and G. barbadense.
Keywords: Amino acid sequence similarity; Disease resistance; Gene structure; Gossypium species; NBS-encoding gene.
Figures
Similar articles
-
Genome-wide characterization and expression analysis of the aldehyde dehydrogenase (ALDH) gene superfamily under abiotic stresses in cotton.Gene. 2017 Sep 10;628:230-245. doi: 10.1016/j.gene.2017.07.034. Epub 2017 Jul 12. Gene. 2017. PMID: 28711668
-
Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species.Plant Biol (Stuttg). 2017 May;19(3):484-493. doi: 10.1111/plb.12536. Epub 2017 Feb 2. Plant Biol (Stuttg). 2017. PMID: 28008701
-
Comparative analysis of resistance gene analogues encoding NBS-LRR domains in cotton.J Sci Food Agric. 2016 Jan 30;96(2):530-8. doi: 10.1002/jsfa.7120. Epub 2015 Mar 3. J Sci Food Agric. 2016. PMID: 25640313
-
Identification of BR biosynthesis genes in cotton reveals that GhCPD-3 restores BR biosynthesis and mediates plant growth and development.Planta. 2021 Sep 17;254(4):75. doi: 10.1007/s00425-021-03727-9. Planta. 2021. PMID: 34533620 Review.
-
Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton.Int J Biol Macromol. 2021 Dec 1;192:1058-1074. doi: 10.1016/j.ijbiomac.2021.10.038. Epub 2021 Oct 14. Int J Biol Macromol. 2021. PMID: 34656543 Review.
Cited by
-
Global whole-genome comparison and analysis to classify subpopulations and identify resistance genes in weedy rice relevant for improving crops.Front Plant Sci. 2023 Jan 10;13:1089445. doi: 10.3389/fpls.2022.1089445. eCollection 2022. Front Plant Sci. 2023. PMID: 36704170 Free PMC article.
-
Identification of circularRNAs and their targets in Gossypium under Verticillium wilt stress based on RNA-seq.PeerJ. 2018 Mar 16;6:e4500. doi: 10.7717/peerj.4500. eCollection 2018. PeerJ. 2018. PMID: 29576969 Free PMC article.
-
Cotton CC-NBS-LRR Gene GbCNL130 Confers Resistance to Verticillium Wilt Across Different Species.Front Plant Sci. 2021 Sep 8;12:695691. doi: 10.3389/fpls.2021.695691. eCollection 2021. Front Plant Sci. 2021. PMID: 34567025 Free PMC article.
-
Heterologous Expression of the Cotton NBS-LRR Gene GbaNA1 Enhances Verticillium Wilt Resistance in Arabidopsis.Front Plant Sci. 2018 Feb 6;9:119. doi: 10.3389/fpls.2018.00119. eCollection 2018. Front Plant Sci. 2018. PMID: 29467784 Free PMC article.
-
Nematode-resistance loci in upland cotton genomes are associated with structural differences.G3 (Bethesda). 2024 Sep 4;14(9):jkae140. doi: 10.1093/g3journal/jkae140. G3 (Bethesda). 2024. PMID: 38934790 Free PMC article.
References
-
- Wendel J, Brubaker C, Alvarez I, Cronn R. In: Genetics and genomics of cotton. Paterson AH, editor. New York: Springer; 2009. pp. 3–22.
-
- Pegg G, Brady B. Verticillium wilts 552. New York: CABI; 2002.
-
- Khadi BM, Santhy V. In: Cotton: biotechnological advances. Zehr UB, editor. New York: Springer; 2010. pp. 15–44.
-
- Du W, Du X, Ma S. Progress of inheritance and molecular biology of Verticillium wilt resistance in cotton. Cotton Sci (China) 2002;14:311–317.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

