Determining the presence of asthma-related molecules and salivary contamination in exhaled breath condensate
- PMID: 28403875
- PMCID: PMC5389118
- DOI: 10.1186/s12931-017-0538-5
Determining the presence of asthma-related molecules and salivary contamination in exhaled breath condensate
Abstract
Background: Researchers investigating lung diseases, such as asthma, have questioned whether certain compounds previously reported in exhaled breath condensate (EBC) originate from saliva contamination. Moreover, despite its increasing use in 'omics profiling studies, the constituents of EBC remain largely uncharacterized. The present study aims to define the usefulness of EBC in investigating lung disease by comparing EBC, saliva, and saliva-contaminated EBC using targeted and untargeted mass spectrometry and the potential of metabolite loss from adsorption to EBC sample collection tubes.
Methods: Liquid chromatography mass spectrometry (LC-MS) was used to analyze samples from 133 individuals from three different cohorts. Levels of amino acids and eicosanoids, two classes of molecules previously reported in EBC and saliva, were measured using targeted LC-MS. Cohort 1 was used to examine contamination of EBC by saliva. Samples from Cohort 1 consisted of clean EBC, saliva-contaminated EBC, and clean saliva from 13 healthy volunteers; samples were analyzed using untargeted LC-MS. Cohort 2 was used to compare eicosanoid levels from matched EBC and saliva collected from 107 asthmatic subjects. Samples were analyzed using both targeted and untargeted LC-MS. Cohort 3 samples consisted of clean-EBC collected from 13 subjects, including smokers and non-smokers, and were used to independently confirm findings; samples were analyzed using targeted LC-MS, untargeted LC-MS, and proteomics. In addition to human samples, an in-house developed nebulizing system was used to determine the potential for EBC samples to be contaminated by saliva.
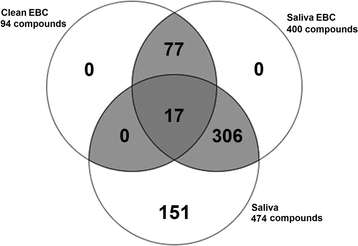
Results: Out of the 400 metabolites detected in both EBC and saliva, 77 were specific to EBC; however, EBC samples were concentrated 20-fold to achieve this level of sensitivity. Amino acid concentrations ranged from 196 pg/mL - 4 μg/mL (clean EBC), 1.98 ng/mL - 6 μg/mL (saliva-contaminated EBC), and 13.84 ng/mL - 1256 mg/mL (saliva). Eicosanoid concentration ranges were an order of magnitude lower; 10 pg/mL - 76.5 ng/mL (clean EBC), 10 pg/mL - 898 ng/mL (saliva-contaminated EBC), and 2.54 ng/mL - 272.9 mg/mL (saliva). Although the sample size of the replication cohort (Cohort 3) did not allow for statistical comparisons, two proteins and 19 eicosanoids were detected in smoker vs. non-smoker clean-EBC.
Conclusions: We conclude that metabolites are present and detectable in EBC using LC-MS; however, a large starting volume of sample is required.
Keywords: Amino acids; Asthma; EBC; Eicosanoids; LC-MS; Leukotriene; Lung; Metabolomics; Proteomics; Saliva.
Figures




References
-
- Brzozowska A, Majak P, Jerzyńska J, Smejda K, Bobrowska-Korzeniowska M, Stelmach W, et al. Exhaled nitric oxide correlates with IL-2, MCP-1, PDGF-BB and TIMP-2 in exhaled breath condensate of children with refractory asthma. Adv Dermatol Allergol. 2015;32(2):107–13. doi: 10.5114/pdia.2014.40953. - DOI - PMC - PubMed
-
- Esther CR, Jr, Boysen G, Olsen BM, Collins LB, Ghio AJ, Swenberg JW, et al. Mass spectrometric analysis of biomarkers and dilution markers in exhaled breath condensate reveals elevated purines in asthma and cystic fibrosis. Am J Physiol Lung Cell Mol Physiol. 2009;296(6):L987–L93. doi: 10.1152/ajplung.90512.2008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

