DC - SIGNR by influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4 in gastric cancer liver metastasis
- PMID: 28403883
- PMCID: PMC5390362
- DOI: 10.1186/s12943-017-0639-2
DC - SIGNR by influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4 in gastric cancer liver metastasis
Abstract
Background: Profiling evidences of selectin demonstrate that they play an crucial role in cancer progression and metastasis. However, DC-SIGNR as a family member of selectin participates in gastric cancer liver metastasis remains unknown.
Methods: The serum level of DC-SIGNR was evaluated in gastric cancer patients by ELISA. Manipulation DC-SIGNR expression in BGC823 and SGC7901 cell lines was mediated by lentivirus. Investigation the biological effects of DC-SIGNR were verified by MTT, wounding and transwell in vitro and experiments on animals to confirm gastric cancer liver metastasis by IVIS. Insights of the mechanism were employed microarray and bioinformatic analysis. Further to confirm the results were conducted by qRT-PCR, western blot and by flow cytometry.
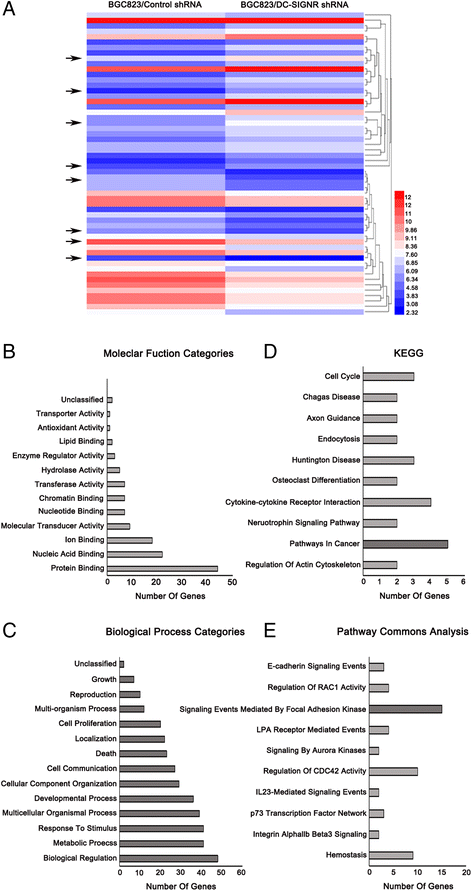
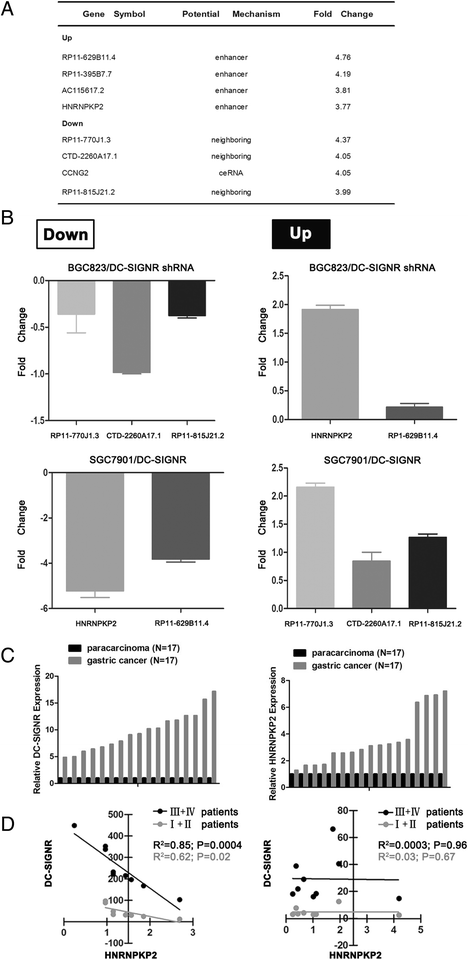
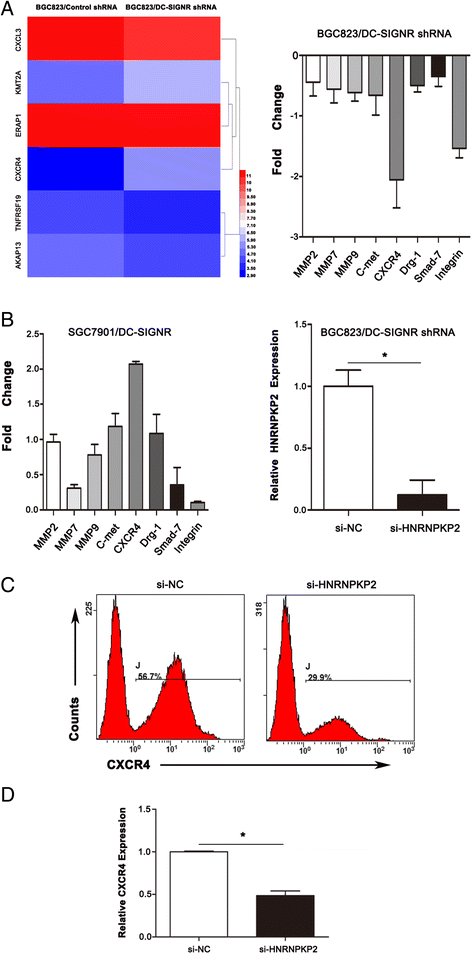
Results: DC-SIGNR serum level was significantly increased in gastric cancer patients compared with healthy group. Additionally, DC-SIGNR level was associated with an advanced pathological stage in gastric cancer patients. DC-SIGNR knockdown inhibited the proliferation, migration and invasion of gastric cancer cells in vitro and suppressed the liver metastasis in vivo. While, DC-SIGNR overexpression promoted cell proliferation, migration and invasion. In mechanism, HNRNPKP2 as a lncRNA was upregulated after DC-SIGNR knockdown. Importantly, STAT5A promoted HNRNPKP2 expression after knockdown DC-SIGNR. Furthermore after HNRNPKP2 depletion, the downstream target gene CXCR4 was downregulated.
Conclusions: DC-SIGNR promoted gastric cancer liver metastasis mediated with HNRNPKP2 which expression was regulated by STAT5A. And HNRNPKP2 decreased the expression of downstream target gene CXCR4. These findings indicated potential therapeutic candidates for gastric cancer liver metastasis.
Keywords: CXCR4; DC-SIGNR; Gastric cancer; Liver metastasis; STAT5A; lncRNA HNRNPKP2.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

