H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity
- PMID: 28403905
- PMCID: PMC5389094
- DOI: 10.1186/s13059-017-1197-z
H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity
Abstract
Background: Polycomb group complexes PRC1 and PRC2 repress gene expression at the chromatin level in eukaryotes. The classic recruitment model of Polycomb group complexes in which PRC2-mediated H3K27 trimethylation recruits PRC1 for H2A monoubiquitination was recently challenged by data showing that PRC1 activity can also recruit PRC2. However, the prevalence of these two mechanisms is unknown, especially in plants as H2AK121ub marks were examined at only a handful of Polycomb group targets.
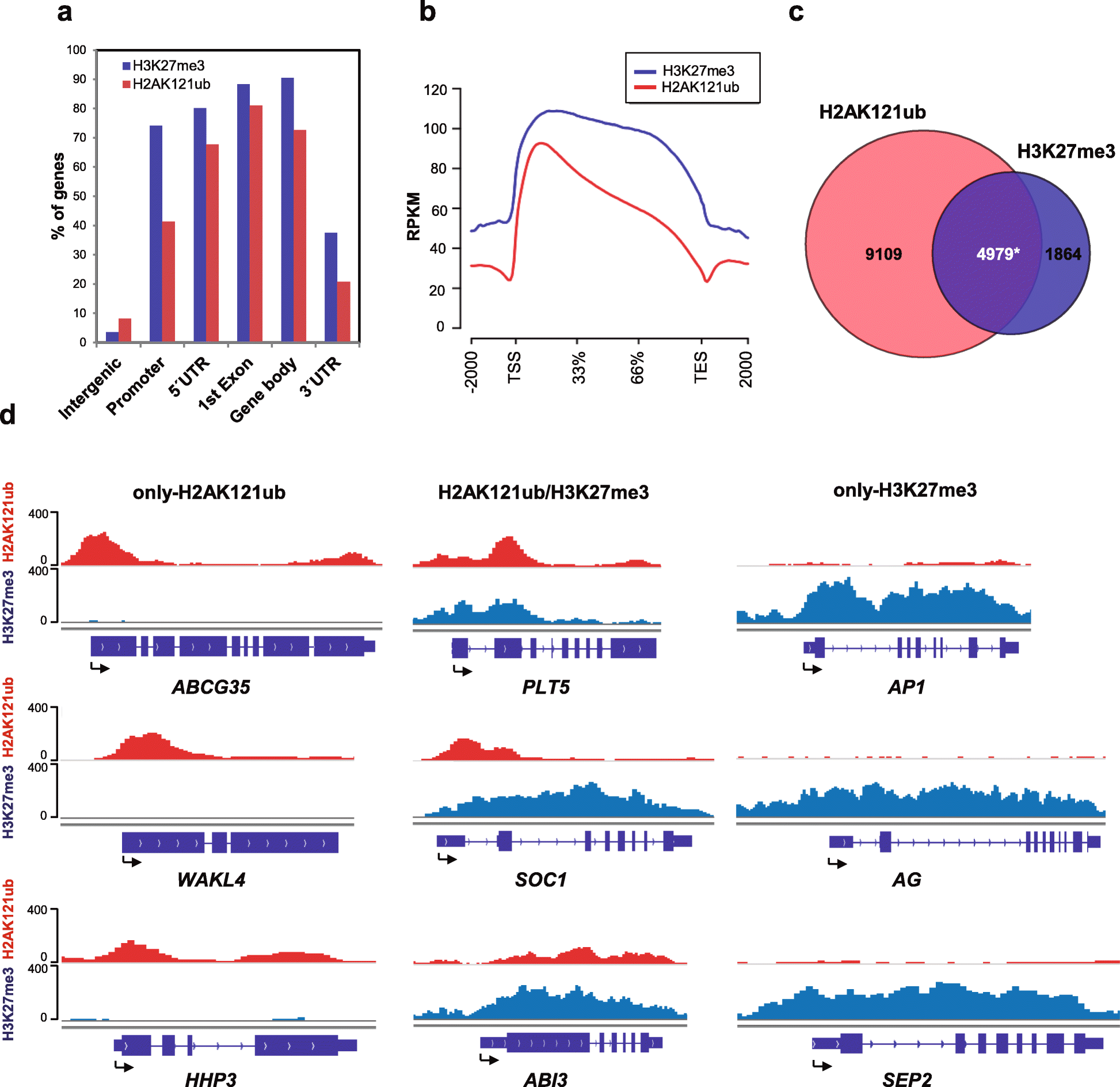
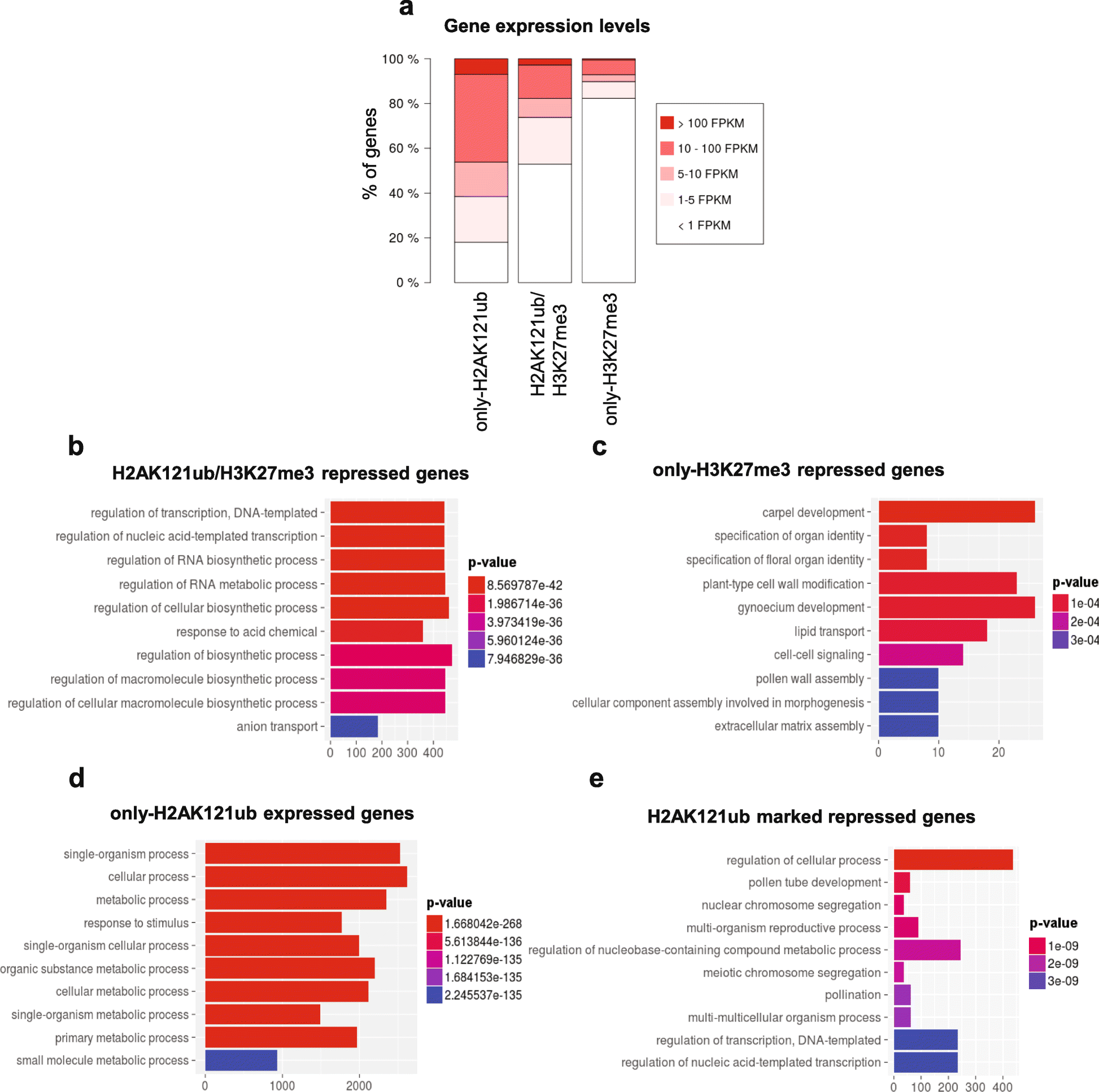
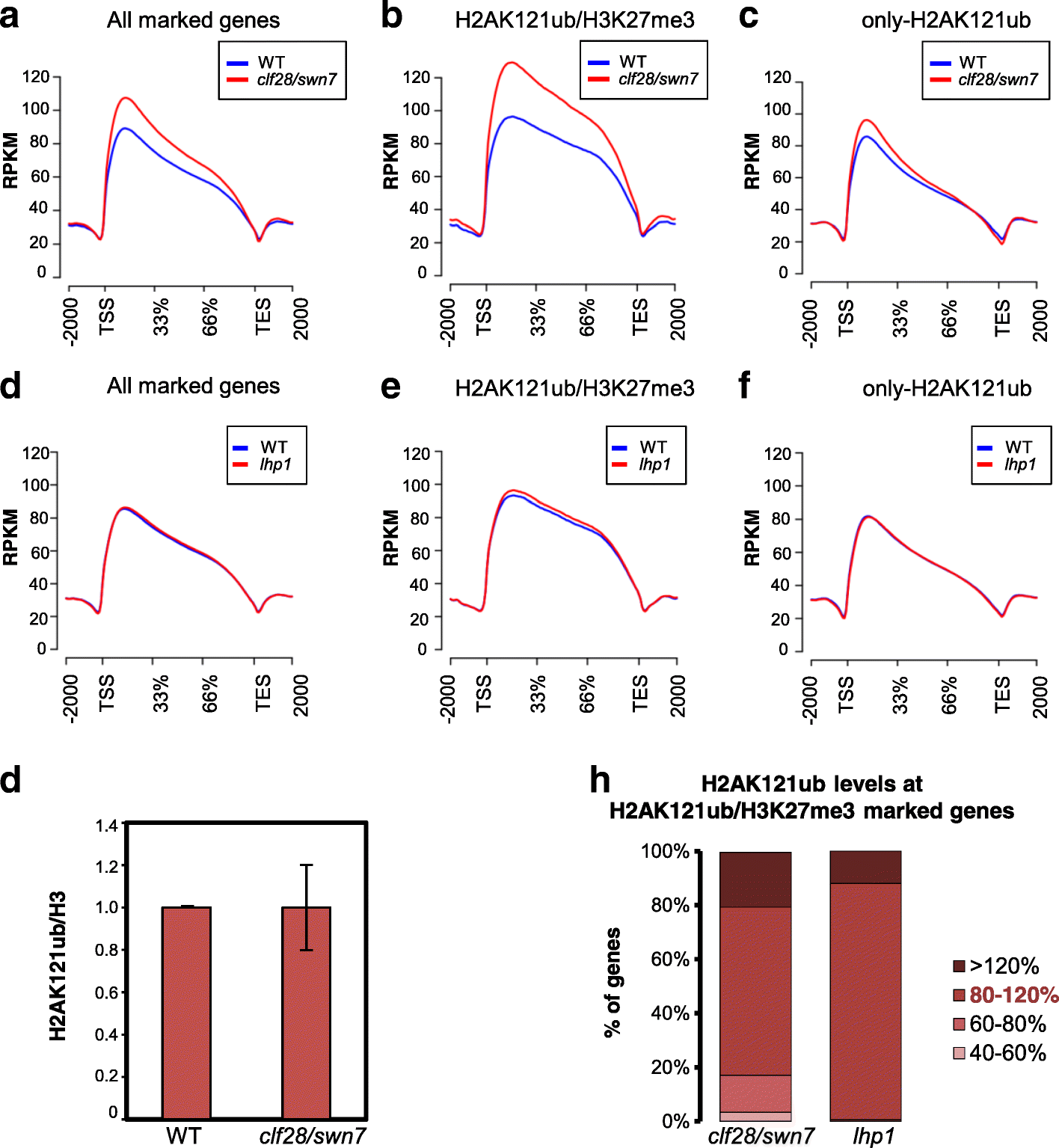
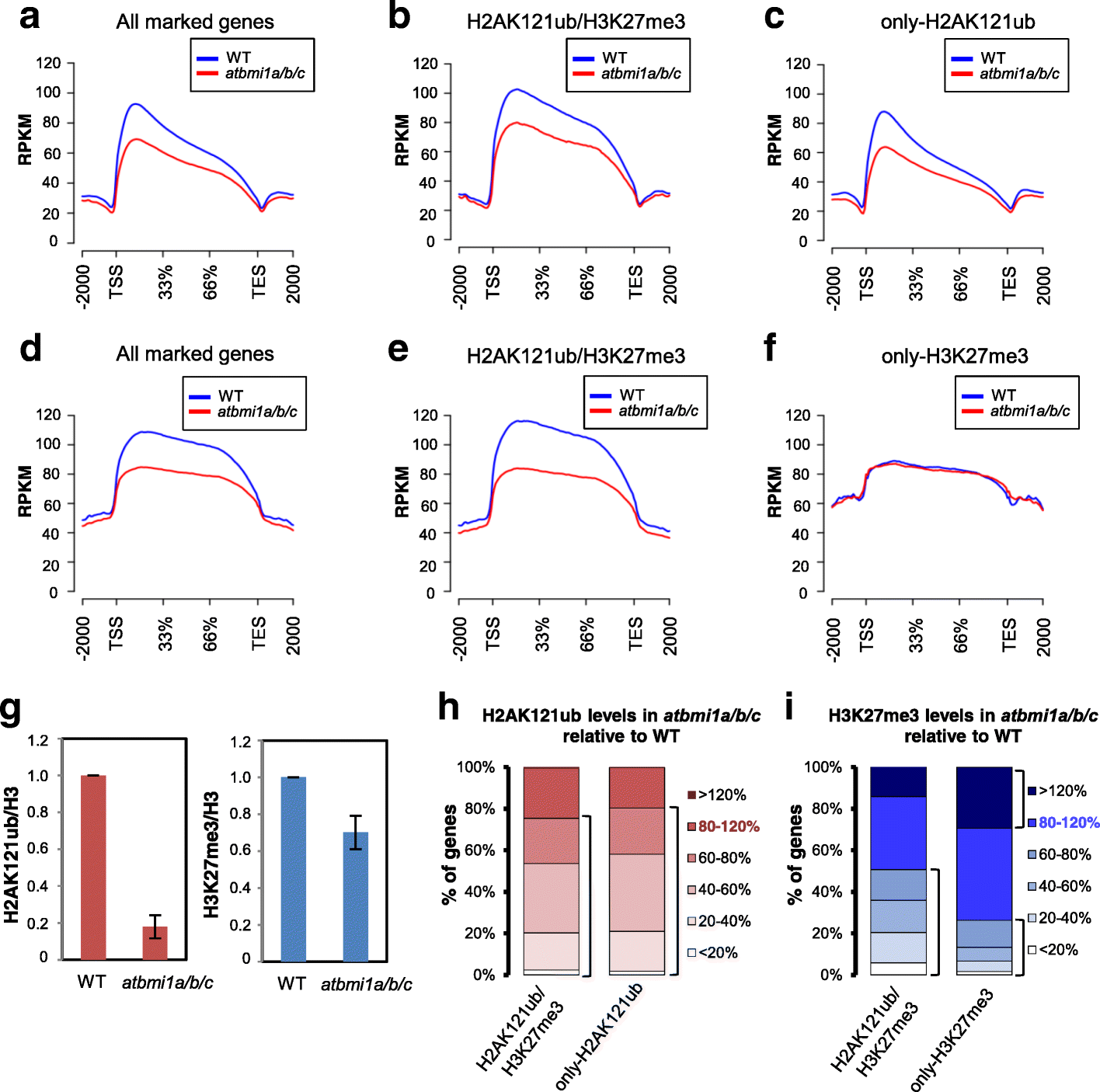
Results: By using genome-wide analyses, we show that H2AK121ub marks are surprisingly widespread in Arabidopsis thaliana, often co-localizing with H3K27me3 but also occupying a set of transcriptionally active genes devoid of H3K27me3. Furthermore, by profiling H2AK121ub and H3K27me3 marks in atbmi1a/b/c, clf/swn, and lhp1 mutants we found that PRC2 activity is not required for H2AK121ub marking at most genes. In contrast, loss of AtBMI1 function impacts the incorporation of H3K27me3 marks at most Polycomb group targets.
Conclusions: Our findings show the relationship between H2AK121ub and H3K27me3 marks across the A. thaliana genome and unveil that ubiquitination by PRC1 is largely independent of PRC2 activity in plants, while the inverse is true for H3K27 trimethylation.
Keywords: Arabidopsis thaliana; H2AK121ub; H3K27me3; PRC1; PRC2; Polycomb group repression mechanism.
Figures

Similar articles
-
PRC1 is taking the lead in PcG repression.Plant J. 2015 Jul;83(1):110-20. doi: 10.1111/tpj.12818. Epub 2015 Mar 20. Plant J. 2015. PMID: 25754661 Review.
-
Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2017 May 2;114(18):4833-4838. doi: 10.1073/pnas.1620955114. Epub 2017 Apr 20. Proc Natl Acad Sci U S A. 2017. PMID: 28428341 Free PMC article.
-
The Arabidopsis Polycomb Repressive Complex 1 (PRC1) Components AtBMI1A, B, and C Impact Gene Networks throughout All Stages of Plant Development.Plant Physiol. 2017 Jan;173(1):627-641. doi: 10.1104/pp.16.01259. Epub 2016 Nov 9. Plant Physiol. 2017. PMID: 27837089 Free PMC article.
-
The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis.Nucleic Acids Res. 2021 Jan 11;49(1):98-113. doi: 10.1093/nar/gkaa1129. Nucleic Acids Res. 2021. PMID: 33270882 Free PMC article.
-
Chromatin modulation and gene regulation in plants: insight about PRC1 function.Biochem Soc Trans. 2018 Aug 20;46(4):957-966. doi: 10.1042/BST20170576. Epub 2018 Jul 31. Biochem Soc Trans. 2018. PMID: 30065110 Review.
Cited by
-
Removal of H2Aub1 by ubiquitin-specific proteases 12 and 13 is required for stable Polycomb-mediated gene repression in Arabidopsis.Genome Biol. 2020 Jun 16;21(1):144. doi: 10.1186/s13059-020-02062-8. Genome Biol. 2020. PMID: 32546254 Free PMC article.
-
H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos.Nat Genet. 2021 Apr;53(4):539-550. doi: 10.1038/s41588-021-00820-3. Epub 2021 Apr 5. Nat Genet. 2021. PMID: 33821003
-
The Arabidopsis Nodulin Homeobox Factor AtNDX Interacts with AtRING1A/B and Negatively Regulates Abscisic Acid Signaling.Plant Cell. 2020 Mar;32(3):703-721. doi: 10.1105/tpc.19.00604. Epub 2020 Jan 9. Plant Cell. 2020. PMID: 31919300 Free PMC article.
-
Alternative silencing states of transposable elements in Arabidopsis associated with H3K27me3.Genome Biol. 2025 Jan 20;26(1):11. doi: 10.1186/s13059-024-03466-6. Genome Biol. 2025. PMID: 39833858 Free PMC article.
-
Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in H3K4me3 independently from histone H2B monoubiquitination.Genome Biol. 2019 May 21;20(1):100. doi: 10.1186/s13059-019-1705-4. Genome Biol. 2019. PMID: 31113491 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

