An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks
- PMID: 28403906
- PMCID: PMC5389000
- DOI: 10.1186/s12918-017-0420-6
An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks
Abstract
Background: Weighted Gene Co-expression Network Analysis (WGCNA) is a widely used R software package for the generation of gene co-expression networks (GCN). WGCNA generates both a GCN and a derived partitioning of clusters of genes (modules). We propose k-means clustering as an additional processing step to conventional WGCNA, which we have implemented in the R package km2gcn (k-means to gene co-expression network, https://github.com/juanbot/km2gcn ).
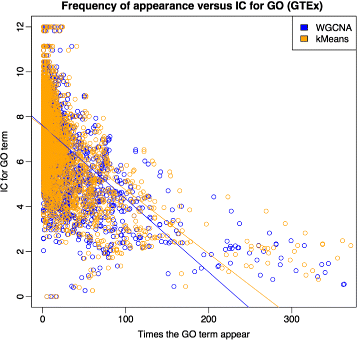
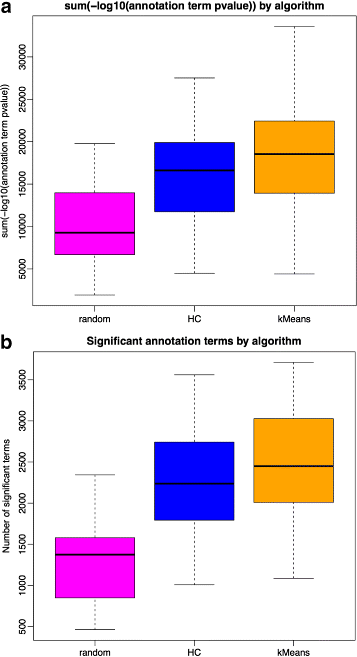
Results: We assessed our method on networks created from UKBEC data (10 different human brain tissues), on networks created from GTEx data (42 human tissues, including 13 brain tissues), and on simulated networks derived from GTEx data. We observed substantially improved module properties, including: (1) few or zero misplaced genes; (2) increased counts of replicable clusters in alternate tissues (x3.1 on average); (3) improved enrichment of Gene Ontology terms (seen in 48/52 GCNs) (4) improved cell type enrichment signals (seen in 21/23 brain GCNs); and (5) more accurate partitions in simulated data according to a range of similarity indices.
Conclusions: The results obtained from our investigations indicate that our k-means method, applied as an adjunct to standard WGCNA, results in better network partitions. These improved partitions enable more fruitful downstream analyses, as gene modules are more biologically meaningful.
Keywords: Assessment of better gene clusters on bulk tissue; Gene co-expression networks on brain; K-means applied to WGCNA.
Figures

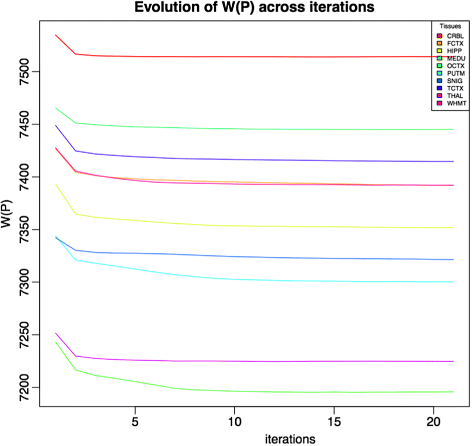
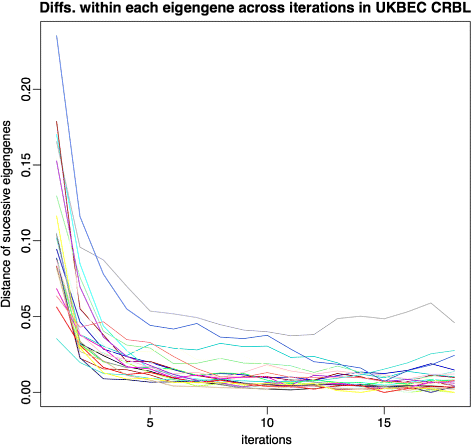
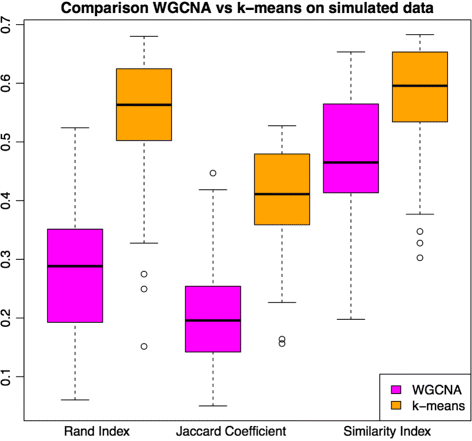
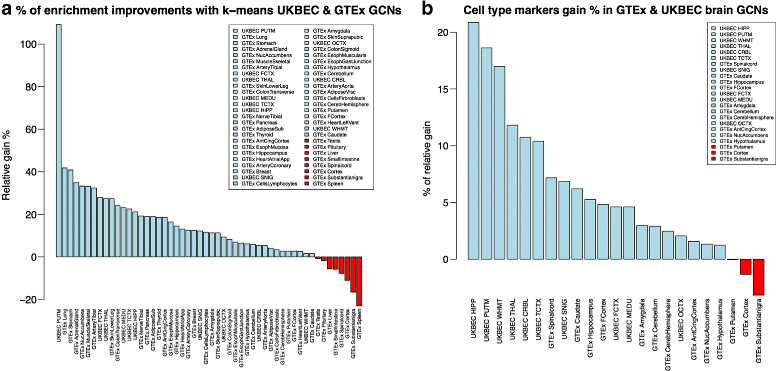
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

