Scaling of cytoskeletal organization with cell size in Drosophila
- PMID: 28404752
- PMCID: PMC5449150
- DOI: 10.1091/mbc.E16-10-0691
Scaling of cytoskeletal organization with cell size in Drosophila
Abstract
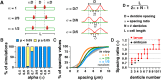
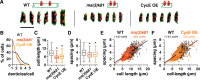
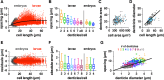
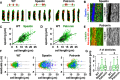
Spatially organized macromolecular complexes are essential for cell and tissue function, but the mechanisms that organize micron-scale structures within cells are not well understood. Microtubule-based structures such as mitotic spindles scale with cell size, but less is known about the scaling of actin structures within cells. Actin-rich denticle precursors cover the ventral surface of the Drosophila embryo and larva and provide templates for cuticular structures involved in larval locomotion. Using quantitative imaging and statistical modeling, we demonstrate that denticle number and spacing scale with cell length over a wide range of cell sizes in embryos and larvae. Denticle number and spacing are reduced under space-limited conditions, and both features robustly scale over a 10-fold increase in cell length during larval growth. We show that the relationship between cell length and denticle spacing can be recapitulated by specific mathematical equations in embryos and larvae and that accurate denticle spacing requires an intact microtubule network and the microtubule minus end-binding protein, Patronin. These results identify a novel mechanism of micro-tubule-dependent actin scaling that maintains precise patterns of actin organization during tissue growth.
© 2017 Spencer et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures



References
-
- Alexandre C, Lecourtois M, Vincent JP. Wingless and Hedgehog pattern Drosophila denticle belts by regulating the production of short-range signals. Development. 1999;126:5689–5698. - PubMed
-
- Atwood HL, Govind CK, Wu CF. Differential ultrastructure of synaptic terminals on ventral longitudinal abdominal muscles in Drosophila larvae. J Neurobiol. 1993;24:1008–1024. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

