Dynamics of the Interaction between Cotton Bollworm Helicoverpa armigera and Nucleopolyhedrovirus as Revealed by Integrated Transcriptomic and Proteomic Analyses
- PMID: 28404795
- PMCID: PMC5461534
- DOI: 10.1074/mcp.M116.062547
Dynamics of the Interaction between Cotton Bollworm Helicoverpa armigera and Nucleopolyhedrovirus as Revealed by Integrated Transcriptomic and Proteomic Analyses
Abstract
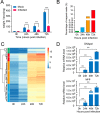
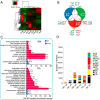
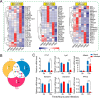
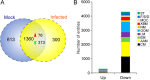
Over the past decades, Helicoverpa armigera nucleopolyhedrovirus (HearNPV) has been widely used for biocontrol of cotton bollworm, which is one of the most destructive pest insects in agriculture worldwide. However, the molecular mechanism underlying the interaction between HearNPV and host insects remains poorly understood. In this study, high-throughput RNA-sequencing was integrated with label-free quantitative proteomics analysis to examine the dynamics of gene expression in the fat body of H. armigera larvae in response to challenge with HearNPV. RNA sequencing-based transcriptomic analysis indicated that host gene expression was substantially altered, yielding 3,850 differentially expressed genes (DEGs), whereas no global transcriptional shut-off effects were observed in the fat body. Among the DEGs, 60 immunity-related genes were down-regulated after baculovirus infection, a finding that was consistent with the results of quantitative real-time RT-PCR. Gene ontology and functional classification demonstrated that the majority of down-regulated genes were enriched in gene cohorts involved in energy, carbohydrate, and amino acid metabolic pathways. Proteomics analysis identified differentially expressed proteins in the fat body, among which 76 were up-regulated, whereas 373 were significantly down-regulated upon infection. The down-regulated proteins are involved in metabolic pathways such as energy metabolism, carbohydrate metabolism (CM), and amino acid metabolism, in agreement with the RNA-sequence data. Furthermore, correlation analysis suggested a strong association between the mRNA level and protein abundance in the H. armigera fat body. More importantly, the predicted gene interaction network indicated that a large subset of metabolic networks was significantly negatively regulated by viral infection, including CM-related enzymes such as aldolase, enolase, malate dehydrogenase, and triose-phosphate isomerase. Taken together, transcriptomic data combined with proteomic data elucidated that baculovirus established systemic infection of host larvae and manipulated the host mainly by suppressing the host immune response and down-regulating metabolism to allow viral self-replication and proliferation. Therefore, this study provided important insights into the mechanism of host-baculovirus interaction.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
High throughput profiling of the cotton bollworm Helicoverpa armigera immunotranscriptome during the fungal and bacterial infections.BMC Genomics. 2015 Apr 18;16(1):321. doi: 10.1186/s12864-015-1509-1. BMC Genomics. 2015. PMID: 26001831 Free PMC article.
-
Digital gene expression analysis of Helicoverpa armigera in the early stage of infection with Helicoverpa armigera nucleopolyhedrovirus.J Invertebr Pathol. 2015 Nov;132:66-76. doi: 10.1016/j.jip.2015.08.008. Epub 2015 Aug 19. J Invertebr Pathol. 2015. PMID: 26296928
-
A Novel Neurotoxin Gene ar1b Recombination Enhances the Efficiency of Helicoverpa armigera Nucleopolyhedrovirus as a Pesticide by Inhibiting the Host Larvae Ability to Feed and Grow.PLoS One. 2015 Aug 21;10(8):e0135279. doi: 10.1371/journal.pone.0135279. eCollection 2015. PLoS One. 2015. PMID: 26296090 Free PMC article.
-
Transcriptomic analysis of interactions between Hyphantria cunea larvae and nucleopolyhedrovirus.Pest Manag Sci. 2019 Apr;75(4):1024-1033. doi: 10.1002/ps.5212. Epub 2018 Nov 7. Pest Manag Sci. 2019. PMID: 30230189
-
Baculovirus induced transcripts in hemocytes from the larvae of Heliothis virescens.Viruses. 2011 Nov;3(11):2047-64. doi: 10.3390/v3112047. Epub 2011 Oct 28. Viruses. 2011. PMID: 22163334 Free PMC article. Review.
Cited by
-
Dual roles and evolutionary implications of P26/poxin in antagonizing intracellular cGAS-STING and extracellular melanization immunity.Nat Commun. 2022 Nov 14;13(1):6934. doi: 10.1038/s41467-022-34761-0. Nat Commun. 2022. PMID: 36376305 Free PMC article.
-
Professor Hu Zhihong: The New President of the Society for Invertebrate Pathology.Virol Sin. 2018 Oct;33(5):383-384. doi: 10.1007/s12250-018-0060-z. Epub 2018 Oct 23. Virol Sin. 2018. PMID: 30353316 Free PMC article. No abstract available.
-
Transcriptomics and interactomics during the primary infection of an SfNPV baculovirus on Spodoptera frugiperda larvae.Front Cell Infect Microbiol. 2023 Nov 21;13:1291433. doi: 10.3389/fcimb.2023.1291433. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38076451 Free PMC article.
-
Identification of a Conserved Prophenoloxidase Activation Pathway in Cotton Bollworm Helicoverpa armigera.Front Immunol. 2020 May 5;11:785. doi: 10.3389/fimmu.2020.00785. eCollection 2020. Front Immunol. 2020. PMID: 32431706 Free PMC article.
-
Population structure of Cydia pomonella granulovirus isolates revealed by quantitative analysis of genetic variation.Virus Evol. 2020 Sep 29;7(1):veaa073. doi: 10.1093/ve/veaa073. eCollection 2021 Jan. Virus Evol. 2020. PMID: 33505705 Free PMC article.
References
-
- Jehle J. A., Lange M., Wang H., Hu Z., Wang Y., and Hauschild R. (2006) Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 346, 180–193 - PubMed
-
- Wu K. M., Lu Y. H., Feng H. Q., Jiang Y. Y., and Zhao J. Z. (2008) Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321, 1676–1678 - PubMed
-
- Szewczyk B., Hoyos-Carvajal L., Paluszek M., Skrzecz I., and Lobo de Souza M. (2006) Baculoviruses–re-emerging biopesticides. Biotechnol. Adv. 24, 143–160 - PubMed
-
- Sun X., Wang H., Sun X., Chen X., Peng C., Pan D., Jehle J. A., van der Werf W., Vlak J. M., and Hu Z. (2004) Biological activity and field efficacy of a genetically modified Helicoverpa armigera single-nucleocapsid nucleopolyhedrovirus expressing an insect-selective toxin from a chimeric promoter. Biol. Control 29, 124–137
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

