Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease
- PMID: 28404864
- PMCID: PMC5497219
- DOI: 10.1126/scitranslmed.aai9321
Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease
Abstract
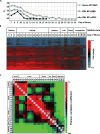
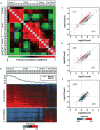
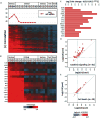
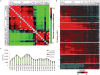
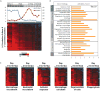
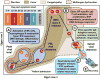
The 2013-2015 outbreak of Ebola virus disease in Guinea, Liberia, and Sierra Leone was unprecedented in the number of documented cases, but there have been few published reports on immune responses in clinical cases and their relationships with the course of illness and severity of Ebola virus disease. Symptoms of Ebola virus disease can include severe headache, myalgia, asthenia, fever, fatigue, diarrhea, vomiting, abdominal pain, and hemorrhage. Although experimental treatments are in development, there are no current U.S. Food and Drug Administration-approved vaccines or therapies. We report a detailed study of host gene expression as measured by microarray in daily peripheral blood samples collected from a patient with severe Ebola virus disease. This individual was provided with supportive care without experimental therapies at the National Institutes of Health Clinical Center from before onset of critical illness to recovery. Pearson analysis of daily gene expression signatures revealed marked gene expression changes in peripheral blood leukocytes that correlated with changes in serum and peripheral blood leukocytes, viral load, antibody responses, coagulopathy, multiple organ dysfunction, and then recovery. This study revealed marked shifts in immune and antiviral responses that preceded changes in medical condition, indicating that clearance of replicating Ebola virus from peripheral blood leukocytes is likely important for systemic viral clearance.
Copyright © 2017, American Association for the Advancement of Science.
Figures


References
-
- World Health Organization. Ebola Situation Report—30 March 2016. World Health Organization; 2016.
-
- Rasmussen AL. Host factors in Ebola infection. Annu Rev Genomics Hum Genet. 2016;17:333–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

