Rad51-mediated double-strand break repair and mismatch correction of divergent substrates
- PMID: 28405019
- PMCID: PMC5544500
- DOI: 10.1038/nature22046
Rad51-mediated double-strand break repair and mismatch correction of divergent substrates
Abstract
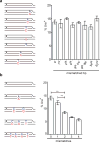
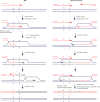
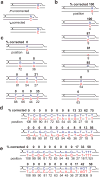
The Rad51 (also known as RecA) family of recombinases executes the critical step in homologous recombination: the search for homologous DNA to serve as a template during the repair of DNA double-strand breaks (DSBs). Although budding yeast Rad51 has been extensively characterized in vitro, the stringency of its search and sensitivity to mismatched sequences in vivo remain poorly defined. Here, in Saccharomyces cerevisiae, we analysed Rad51-dependent break-induced replication in which the invading DSB end and its donor template share a 108-base-pair homology region and the donor carries different densities of single-base-pair mismatches. With every eighth base pair mismatched, repair was about 14% of that of completely homologous sequences. With every sixth base pair mismatched, repair was still more than 5%. Thus, completing break-induced replication in vivo overcomes the apparent requirement for at least 6-8 consecutive paired bases that has been inferred from in vitro studies. When recombination occurs without a protruding nonhomologous 3' tail, the mismatch repair protein Msh2 does not discourage homeologous recombination. However, when the DSB end contains a 3' protruding nonhomologous tail, Msh2 promotes the rejection of mismatched substrates. Mismatch correction of strand invasion heteroduplex DNA is strongly polar, favouring correction close to the DSB end. Nearly all mismatch correction depends on the proofreading activity of DNA polymerase-δ, although the repair proteins Msh2, Mlh1 and Exo1 influence the extent of correction.
Conflict of interest statement
Conflict of Interest: None declared
Figures








References
-
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. - PubMed
-
- Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. - PubMed
-
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

