Hepatitis C virus pharmacogenomics in Latin American populations: implications in the era of direct-acting antivirals
- PMID: 28405170
- PMCID: PMC5378445
- DOI: 10.2147/PGPM.S125452
Hepatitis C virus pharmacogenomics in Latin American populations: implications in the era of direct-acting antivirals
Abstract
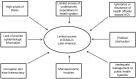
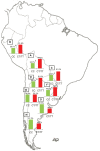
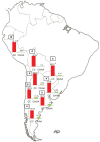
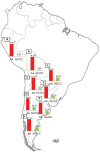
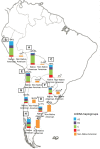
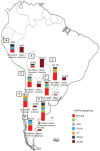
In recent years, great progress has been made in the field of new therapeutic options for hepatitis C virus (HCV) infection. The new direct-acting antiviral agents (DAAs) represent a great hope for millions of chronically infected individuals because their use may lead to excellent cure rates with fewer side effects. In Latin America, the high prevalence of HCV genotype 1 infection and the significant association of Native American ancestry with risk predictive single-nucleotide polymorphisms (SNPs) in IFNL4 and ITPA genes highlight the need to implement new treatment regimens in these populations. However, the universal accessibility to DAAs is still not a reality in the region as their high cost is one of the major, although not the only, limiting factors for their broad implementation. Therefore, under these circumstances, could the assessment of host genetic markers be a useful tool to prioritize DAA treatment until global access to these new drugs can be achieved? This review will summarize the scientific evidences and the potential implications of HCV pharmacogenomics in this rapidly evolving era of anti-HCV drug development.
Keywords: DAAs; Latin America; PEG-IFN/RBV; hepatitis C virus; pharmacogenomics.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures
References
-
- Conteduca V, Sansonno D, Russi S, Pavone F, Dammacco F. Therapy of chronic hepatitis C virus infection in the era of direct-acting and host-targeting antiviral agents. J Infect. 2014;68(1):1–20. - PubMed
-
- Suppiah V, Moldovan M, Ahlenstiel G, et al. Interleukin 28B is associated with response to Hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(1):1100–1104. - PubMed
-
- Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of Interleukin 28B with response to interferon alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–1109. - PubMed
-
- Ge D, Fellay J, Thompson AJ, et al. Genetic variation in interleukin 28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. - PubMed
-
- Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in interleukin 28B is associated with chronic hepatitis C and treatment failure: genome-wide association study. Gastroenterology. 2010;138(4):1338–1345. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

