Targeting the ubiquitin-conjugating enzyme E2D4 for cancer drug discovery-a structure-based approach
- PMID: 28405240
- PMCID: PMC5374094
- DOI: 10.1007/s12154-016-0164-6
Targeting the ubiquitin-conjugating enzyme E2D4 for cancer drug discovery-a structure-based approach
Abstract
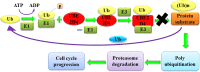
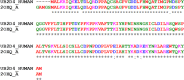
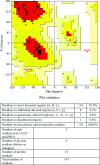
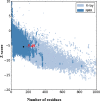
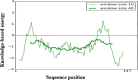
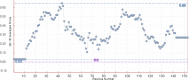
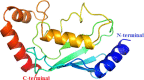
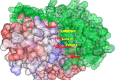
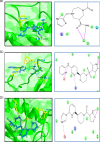
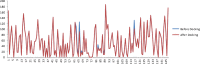
Cancer progression is a global burden. The incidence and mortality now reach 30 million deaths per year. Several pathways of cancer are under investigation for the discovery of effective therapeutics. The present study highlights the structural details of the ubiquitin protein 'Ubiquitin-conjugating enzyme E2D4' (UBE2D4) for the novel lead structure identification in cancer drug discovery process. The evaluation of 3D structure of UBE2D4 was carried out using homology modelling techniques. The optimized structure was validated by standard computational protocols. The active site region of the UBE2D4 was identified using computational tools like CASTp, Q-site Finder and SiteMap. The hydrophobic pocket which is responsible for binding with its natural receptor ubiquitin ligase CHIP (C-terminal of Hsp 70 interacting protein) was identified through protein-protein docking study. Corroborating the results obtained from active site prediction tools and protein-protein docking study, the domain of UBE2D4 which is responsible for cancer cell progression is sorted out for further docking study. Virtual screening with large structural database like CB_Div Set and Asinex BioDesign small molecular structural database was carried out. The obtained new ligand molecules that have shown affinity towards UBE2D4 were considered for ADME prediction studies. The identified new ligand molecules with acceptable parameters of docking, ADME are considered as potent UBE2D4 enzyme inhibitors for cancer therapy.
Keywords: ADME; Active site; Cancer; Homology modelling; Protein-protein docking; Virtual screening.
Conflict of interest statement
Ethical standards
The authors state that no human studies and no animal studies were carried out for this article.
Conflict of interest
The authors declare that they have no conflict of interest.
Figures

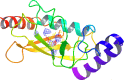



Similar articles
-
Identification of New Lead Molecules Against UBE2NL Enzyme for Cancer Therapy.Appl Biochem Biotechnol. 2017 Aug;182(4):1497-1517. doi: 10.1007/s12010-017-2414-7. Epub 2017 Feb 9. Appl Biochem Biotechnol. 2017. PMID: 28185054
-
Homology modeling and virtual screening studies of FGF-7 protein-a structure-based approach to design new molecules against tumor angiogenesis.J Chem Biol. 2016 Jun 18;9(3):69-78. doi: 10.1007/s12154-016-0152-x. eCollection 2016 Jul. J Chem Biol. 2016. PMID: 27493695 Free PMC article.
-
Identification of Novel Antagonists for Rab38 Protein by Homology Modeling and Virtual Screening.Comb Chem High Throughput Screen. 2016;19(10):875-892. doi: 10.2174/1386207319666161026153237. Comb Chem High Throughput Screen. 2016. PMID: 27784220
-
Free resources to assist structure-based virtual ligand screening experiments.Curr Protein Pept Sci. 2007 Aug;8(4):381-411. doi: 10.2174/138920307781369391. Curr Protein Pept Sci. 2007. PMID: 17696871 Review.
-
Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening.Curr Protein Pept Sci. 2006 Oct;7(5):395-406. doi: 10.2174/138920306778559386. Curr Protein Pept Sci. 2006. PMID: 17073692 Review.
Cited by
-
IGF2BP2-modified UBE2D1 interacts with Smad2/3 to promote the progression of breast cancer.Am J Cancer Res. 2023 Jul 15;13(7):2948-2968. eCollection 2023. Am J Cancer Res. 2023. PMID: 37560007 Free PMC article.
-
The Molecular Basis of Ubiquitin-Conjugating Enzymes (E2s) as a Potential Target for Cancer Therapy.Int J Mol Sci. 2021 Mar 26;22(7):3440. doi: 10.3390/ijms22073440. Int J Mol Sci. 2021. PMID: 33810518 Free PMC article. Review.
-
Computational Approaches in Preclinical Studies on Drug Discovery and Development.Front Chem. 2020 Sep 11;8:726. doi: 10.3389/fchem.2020.00726. eCollection 2020. Front Chem. 2020. PMID: 33062633 Free PMC article. Review.
-
New Insights into the Role of E2s in the Pathogenesis of Diseases: Lessons Learned from UBE2O.Mol Cells. 2018 Mar 31;41(3):168-178. doi: 10.14348/molcells.2018.0008. Epub 2018 Mar 20. Mol Cells. 2018. PMID: 29562734 Free PMC article. Review.
-
In Silico Methods for Identification of Potential Active Sites of Therapeutic Targets.Molecules. 2022 Oct 20;27(20):7103. doi: 10.3390/molecules27207103. Molecules. 2022. PMID: 36296697 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

