Influenza C in Lancaster, UK, in the winter of 2014-2015
- PMID: 28406194
- PMCID: PMC5390268
- DOI: 10.1038/srep46578
Influenza C in Lancaster, UK, in the winter of 2014-2015
Erratum in
-
Erratum: Influenza C in Lancaster, UK, in the winter of 2014-2015.Sci Rep. 2017 May 26;7:46815. doi: 10.1038/srep46815. Sci Rep. 2017. PMID: 28548112 Free PMC article. No abstract available.
Abstract
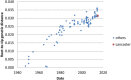
Influenza C is not included in the annual seasonal influenza vaccine, and has historically been regarded as a minor respiratory pathogen. However, recent work has highlighted its potential role as a cause of pneumonia in infants. We performed nasopharyngeal or nasal swabbing and/or serum sampling (n = 148) in Lancaster, UK, over the winter of 2014-2015. Using enzyme-linked immunosorbent assay (ELISA), we obtain seropositivity of 77%. By contrast, only 2 individuals, both asymptomatic adults, were influenza C-positive by polymerase chain reaction (PCR). Deep sequencing of nasopharyngeal samples produced partial sequences for 4 genome segments in one of these patients. Bayesian phylogenetic analysis demonstrated that the influenza C genome from this individual is evolutionarily distant to those sampled in recent years and represents a novel genome constellation, indicating that it may be a product of a decades-old reassortment event. Although we find no evidence that influenza C was a significant respiratory pathogen during the winter of 2014-2015 in Lancaster, we confirm previous observations of seropositivity in the majority of the population. (170 words).
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

