NNAlign: a platform to construct and evaluate artificial neural network models of receptor-ligand interactions
- PMID: 28407117
- PMCID: PMC5570195
- DOI: 10.1093/nar/gkx276
NNAlign: a platform to construct and evaluate artificial neural network models of receptor-ligand interactions
Abstract
Peptides are extensively used to characterize functional or (linear) structural aspects of receptor-ligand interactions in biological systems, e.g. SH2, SH3, PDZ peptide-recognition domains, the MHC membrane receptors and enzymes such as kinases and phosphatases. NNAlign is a method for the identification of such linear motifs in biological sequences. The algorithm aligns the amino acid or nucleotide sequences provided as training set, and generates a model of the sequence motif detected in the data. The webserver allows setting up cross-validation experiments to estimate the performance of the model, as well as evaluations on independent data. Many features of the training sequences can be encoded as input, and the network architecture is highly customizable. The results returned by the server include a graphical representation of the motif identified by the method, performance values and a downloadable model that can be applied to scan protein sequences for occurrence of the motif. While its performance for the characterization of peptide-MHC interactions is widely documented, we extended NNAlign to be applicable to other receptor-ligand systems as well. Version 2.0 supports alignments with insertions and deletions, encoding of receptor pseudo-sequences, and custom alphabets for the training sequences. The server is available at http://www.cbs.dtu.dk/services/NNAlign-2.0.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


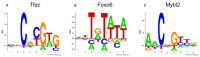
References
-
- Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. - PubMed
-
- Nielsen M., Lundegaard C., Worning P., Hvid C.S., Lamberth K., Buus S., Brunak S., Lund O.. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics. 2004; 20:1388–1397. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

