Designed Spiroketal Protein Modulation
- PMID: 28407400
- PMCID: PMC5435924
- DOI: 10.1002/anie.201612504
Designed Spiroketal Protein Modulation
Abstract
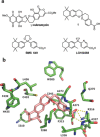
Spiroketals are structural motifs found in many biologically active natural products, which has stimulated considerable efforts toward their synthesis and interest in their use as drug lead compounds. Despite this, the use of spiroketals, and especially bisbenzanulated spiroketals, in a structure-based drug discovery setting has not been convincingly demonstrated. Herein, we report the rational design of a bisbenzannulated spiroketal that potently binds to the retinoid X receptor (RXR) thereby inducing partial co-activator recruitment. We solved the crystal structure of the spiroketal-hRXRα-TIF2 ternary complex, and identified a canonical allosteric mechanism as a possible explanation for the partial agonist behavior of our spiroketal. Our co-crystal structure, the first of a designed spiroketal-protein complex, suggests that spiroketals can be designed to selectively target other nuclear receptor subtypes.
Keywords: drug design; drug discovery; natural products; spiro compounds; structure elucidation.
© 2017 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Figures



References
-
- Zheng Y., Tice C. M., Singh S. B., Bioorg. Med. Chem. Lett. 2014, 24, 3673–3682. - PubMed
-
- Perron F., Albizati K. F., Chem. Rev. 1989, 89, 1617–1661.
-
- Sperry J., Wilson Z. E., Rathwell D. C. K., Brimble M. A., Nat. Prod. Rep. 2010, 27, 1117–1137. - PubMed
-
- Yoneda N., Fukata Y., Asano K., Matsubara S., Angew. Chem. Int. Ed. 2015, 54, 15497–15500; - PubMed
- Angew. Chem. 2015, 127, 15717–15720.
-
- Butler B. B., Manda J. N., Aponick A., Org. Lett. 2015, 17, 1902–1905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

