Clinical Variant Classification: A Comparison of Public Databases and a Commercial Testing Laboratory
- PMID: 28408614
- PMCID: PMC5507641
- DOI: 10.1634/theoncologist.2016-0431
Clinical Variant Classification: A Comparison of Public Databases and a Commercial Testing Laboratory
Abstract
Background: There is a growing move to consult public databases following receipt of a genetic test result from a clinical laboratory; however, the well-documented limitations of these databases call into question how often clinicians will encounter discordant variant classifications that may introduce uncertainty into patient management. Here, we evaluate discordance in BRCA1 and BRCA2 variant classifications between a single commercial testing laboratory and a public database commonly consulted in clinical practice.
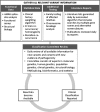
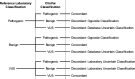
Materials and methods: BRCA1 and BRCA2 variant classifications were obtained from ClinVar and compared with the classifications from a reference laboratory. Full concordance and discordance were determined for variants whose ClinVar entries were of the same pathogenicity (pathogenic, benign, or uncertain). Variants with conflicting ClinVar classifications were considered partially concordant if ≥1 of the listed classifications agreed with the reference laboratory classification.
Results: Four thousand two hundred and fifty unique BRCA1 and BRCA2 variants were available for analysis. Overall, 73.2% of classifications were fully concordant and 12.3% were partially concordant. The remaining 14.5% of variants had discordant classifications, most of which had a definitive classification (pathogenic or benign) from the reference laboratory compared with an uncertain classification in ClinVar (14.0%).
Conclusion: Here, we show that discrepant classifications between a public database and single reference laboratory potentially account for 26.7% of variants in BRCA1 and BRCA2. The time and expertise required of clinicians to research these discordant classifications call into question the practicality of checking all test results against a database and suggest that discordant classifications should be interpreted with these limitations in mind.
Implications for practice: With the increasing use of clinical genetic testing for hereditary cancer risk, accurate variant classification is vital to ensuring appropriate medical management. There is a growing move to consult public databases following receipt of a genetic test result from a clinical laboratory; however, we show that up to 26.7% of variants in BRCA1 and BRCA2 have discordant classifications between ClinVar and a reference laboratory. The findings presented in this paper serve as a note of caution regarding the utility of database consultation.
Keywords: BRCA1; BRCA2; Genetic testing; Public databases; Variant classification.
© AlphaMed Press 2017.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures
Comment in
-
In Reply.Oncologist. 2017 Dec;22(12):1540. doi: 10.1634/theoncologist.2017-0320. Epub 2017 Aug 29. Oncologist. 2017. PMID: 28851761 Free PMC article.
-
ClinVar Is a Critical Resource to Advance Variant Interpretation.Oncologist. 2017 Dec;22(12):1562. doi: 10.1634/theoncologist.2017-0246. Epub 2017 Aug 29. Oncologist. 2017. PMID: 28851762 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous