MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff
- PMID: 28408736
- PMCID: PMC5429825
- DOI: 10.1038/s41598-017-00853-x
MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff
Abstract
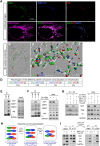
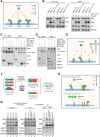
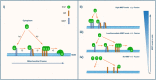
Mitochondrial dynamics is a fundamental cellular process and recruitment of Drp1 to mitochondria is an essential step in mitochondrial fission. Mff and MIEF1/2 (MiD51/49) serve as key receptors for recruitment of Drp1 to mitochondria in mammals. However, if and how these receptors work together in mitochondrial fission is poorly understood. Here we show that MIEFs interact with both Drp1 and Mff on the mitochondrial surface and serve as adaptors linking Drp1 and Mff together in a trimeric Drp1-MIEF-Mff complex. Thus, MIEFs can regulate the interaction between Drp1 and Mff, and also Mff-induced Drp1 accumulation on mitochondria. It is shown that loss of endogenous MIEFs severely impairs these processes. Additionally, in cells depleted of endogenous MIEF1/2, high levels of exogenous MIEFs sequester Drp1 on the mitochondrial surface, resulting in mitochondrial elongation, whereas low-to-moderate levels of MIEFs promote mitochondrial fission, leading to mitochondrial fragmentation. In sum, the data suggest that MIEFs and Mff work coordinately in Drp1-mediated mitochondrial fission and that the level of MIEF1/2 relative to Mff sets the balance between mitochondrial fission and fusion.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

