A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana
- PMID: 28410577
- PMCID: PMC5391559
- DOI: 10.1186/s12870-017-1016-4
A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana
Abstract
Background: Diverse plant pathogens deliver effectors into plant cells to alter host processes. Oomycete pathogen encodes a large number of putative RxLR effectors which are likely to play a role in manipulating plant defense responses. The secretome of Plasmopara viticola (downy mildew of grapevine) contains at least 162 candidate RxLR effectors discovered in our recent studies, but their roles in infection and pathogenicity remain to be determined. Here, we characterize in depth one of the putative RxLR effectors, PvRxLR16, which has been reported to induce cell death in Nicotiana benthamiana in our previous study.
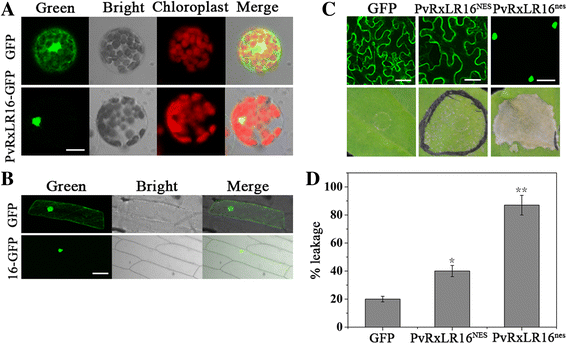
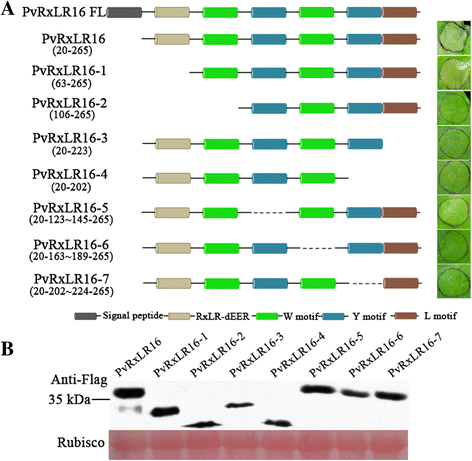
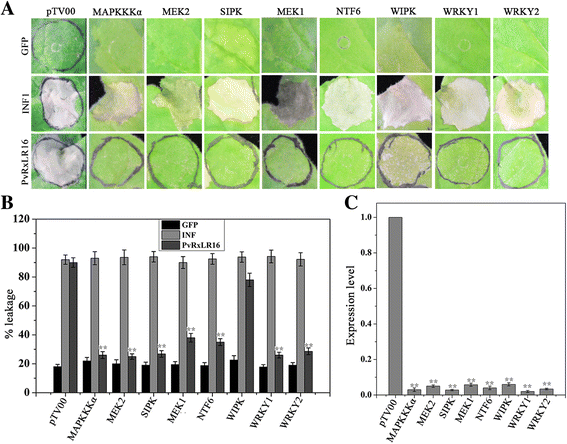
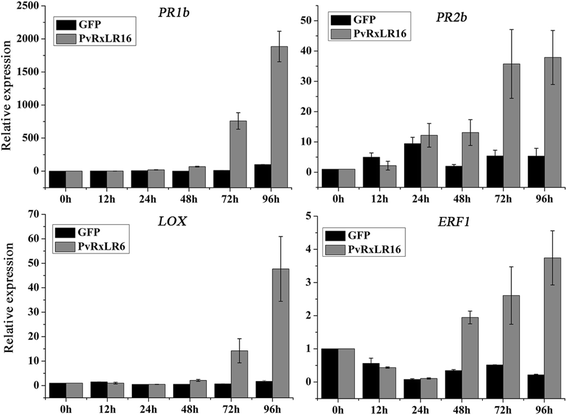
Results: The nuclear localization, W/Y/L motifs, and a putative N-glycosylation site in C-terminal of PvRxLR16 were essential for cell death-inducing activity. Suppressor of G-two allele of Skp1 (SGT1), heat shock protein 90 (HSP90) and required for Mla12 resistance (RAR1), but not somatic embryogenesis receptor-like kinase (SERK3), were required for the cell death response triggered by PvRxLR16 in N. benthamiana. Some mitogen-activated protein kinases and transcription factors were also involved in the perception of PvRxLR16 by N. benthamiana. PvRxLR16 could also significantly enhance plant resistance to Phytophthora capsici and the nuclear localization was required for this ability. However, some other PvRxLR effectors could suppress defense responses and disease resistance induced by PvRxLR16, suggesting that it may not trigger host cell death or immune responses during physiological infection under natural conditions.
Conclusion: These data demonstrate that PvRxLR16 may be recognized by endogenous proteins in nucleus to trigger immune responses in N. benthamiana, which in turn can be suppressed by other PvRxLR effectors.
Keywords: Nicotiana benthamiana cell death; Plasmopara viticola; RxLR effector; grapevine; immune responses.
Figures





Similar articles
-
The Nuclear-Localized RxLR Effector PvAvh74 From Plasmopara viticola Induces Cell Death and Immunity Responses in Nicotiana benthamiana.Front Microbiol. 2019 Jul 10;10:1531. doi: 10.3389/fmicb.2019.01531. eCollection 2019. Front Microbiol. 2019. PMID: 31354650 Free PMC article.
-
Plasmopara viticola effector PvRXLR159 suppresses immune responses in Nicotiana benthamiana.Plant Signal Behav. 2019;14(12):1682220. doi: 10.1080/15592324.2019.1682220. Epub 2019 Oct 24. Plant Signal Behav. 2019. PMID: 31647363 Free PMC article.
-
Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly related plants.Plant J. 2012 Dec;72(6):882-93. doi: 10.1111/j.1365-313X.2012.05079.x. Epub 2012 Oct 26. Plant J. 2012. PMID: 22709376
-
Recent Progress in RXLR Effector Research.Mol Plant Microbe Interact. 2015 Oct;28(10):1063-72. doi: 10.1094/MPMI-01-15-0022-CR. Epub 2015 Oct 2. Mol Plant Microbe Interact. 2015. PMID: 26125490 Review.
-
The pathogenicity of Plasmopara viticola: a review of evolutionary dynamics, infection strategies and effector molecules.BMC Plant Biol. 2024 Apr 24;24(1):327. doi: 10.1186/s12870-024-05037-0. BMC Plant Biol. 2024. PMID: 38658826 Free PMC article. Review.
Cited by
-
Candidate effector proteins from the oomycetes Plasmopara viticola and Phytophthora parasitica share similar predicted structures and induce cell death in Nicotiana species.PLoS One. 2022 Dec 2;17(12):e0278778. doi: 10.1371/journal.pone.0278778. eCollection 2022. PLoS One. 2022. PMID: 36459530 Free PMC article.
-
Functional analysis of the Nep1-like proteins from Plasmopara viticola.Plant Signal Behav. 2022 Dec 31;17(1):2000791. doi: 10.1080/15592324.2021.2000791. Epub 2022 Feb 12. Plant Signal Behav. 2022. PMID: 35152834 Free PMC article.
-
A Putative Effector LtCSEP1 from Lasiodiplodia theobromae Inhibits BAX-Triggered Cell Death and Suppresses Immunity Responses in Nicotiana benthamiana.Plants (Basel). 2022 May 30;11(11):1462. doi: 10.3390/plants11111462. Plants (Basel). 2022. PMID: 35684232 Free PMC article.
-
An RXLR effector secreted by Phytophthora parasitica is a virulence factor and triggers cell death in various plants.Mol Plant Pathol. 2019 Mar;20(3):356-371. doi: 10.1111/mpp.12760. Epub 2018 Nov 22. Mol Plant Pathol. 2019. PMID: 30320960 Free PMC article.
-
Genome-Wide Identification and Analysis of Gene Family of Carbohydrate-Binding Modules in Ustilago crameri.Int J Mol Sci. 2024 Nov 2;25(21):11790. doi: 10.3390/ijms252111790. Int J Mol Sci. 2024. PMID: 39519340 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

