MicroRNA expression profiling of Xp11 renal cell carcinoma
- PMID: 28411178
- PMCID: PMC5628161
- DOI: 10.1016/j.humpath.2017.03.011
MicroRNA expression profiling of Xp11 renal cell carcinoma
Abstract
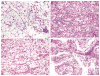
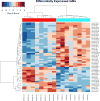
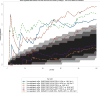
Renal cell carcinomas (RCCs) with Xp11 translocation (Xp11 RCC) constitute a distinctive molecular subtype characterized by chromosomal translocations involving the Xp11.2 locus, resulting in gene fusions between the TFE3 transcription factor with a second gene (usually ASPSCR1, PRCC, NONO, or SFPQ). RCCs with Xp11 translocations comprise up to 1% to 4% of adult cases, frequently displaying papillary architecture with epithelioid clear cells. To better understand the biology of this molecularly distinct tumor subtype, we analyze the microRNA (miRNA) expression profiles of Xp11 RCC compared with normal renal parenchyma using microarray and quantitative reverse-transcription polymerase chain reaction. We further compare Xp11 RCC with other RCC histologic subtypes using publically available data sets, identifying common and distinctive miRNA signatures along with the associated signaling pathways and biological processes. Overall, Xp11 RCC more closely resembles clear cell rather than papillary RCC. Furthermore, among the most differentially expressed miRNAs specific for Xp11 RCC, we identify miR-148a-3p, miR-221-3p, miR-185-5p, miR-196b-5p, and miR-642a-5p to be up-regulated, whereas miR-133b and miR-658 were down-regulated. Finally, Xp11 RCC is most strongly associated with miRNA expression profiles modulating DNA damage responses, cell cycle progression and apoptosis, and the Hedgehog signaling pathway. In summary, we describe here for the first time the miRNA expression profiles of a molecularly distinct type of renal cancer associated with Xp11.2 translocations involving the TFE3 gene. Our results might help understanding the molecular underpinning of Xp11 RCC, assisting in developing targeted treatments for this disease.
Keywords: Analysis of Functional Annotation; Renal cell carcinoma; TFE3 gene fusion; Xp11 translocation; gene regulation; microRNA expression profiling.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures


Comment in
-
MicroRNAs, promising biomarkers in the diagnosis of Xp11 translocation RCC.Hum Pathol. 2017 Oct;68:205-206. doi: 10.1016/j.humpath.2017.06.022. Epub 2017 Aug 12. Hum Pathol. 2017. PMID: 28807734 No abstract available.
-
MicroRNAs, promising biomarkers in the diagnosis of Xp11 translocation RCC-reply.Hum Pathol. 2017 Oct;68:206-207. doi: 10.1016/j.humpath.2017.06.021. Epub 2017 Aug 12. Hum Pathol. 2017. PMID: 28811253 No abstract available.
Similar articles
-
TFE3-Fusion Variant Analysis Defines Specific Clinicopathologic Associations Among Xp11 Translocation Cancers.Am J Surg Pathol. 2016 Jun;40(6):723-37. doi: 10.1097/PAS.0000000000000631. Am J Surg Pathol. 2016. PMID: 26975036 Free PMC article.
-
TMED6-COG8 is a novel molecular marker of TFE3 translocation renal cell carcinoma.Int J Clin Exp Pathol. 2015 Mar 1;8(3):2690-9. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26045774 Free PMC article.
-
[Clinicopathologic and molecular genetic study of renal cell carcinoma occurring in teenagers].Zhonghua Bing Li Xue Za Zhi. 2010 Sep;39(9):582-6. Zhonghua Bing Li Xue Za Zhi. 2010. PMID: 21092583 Chinese.
-
Clinical heterogeneity of Xp11 translocation renal cell carcinoma: impact of fusion subtype, age, and stage.Mod Pathol. 2014 Jun;27(6):875-86. doi: 10.1038/modpathol.2013.208. Epub 2013 Dec 6. Mod Pathol. 2014. PMID: 24309327 Review.
-
RBM10-TFE3 renal cell carcinoma characterised by paracentric inversion with consistent closely split signals in break-apart fluorescence in-situ hybridisation: study of 10 cases and a literature review.Histopathology. 2019 Aug;75(2):254-265. doi: 10.1111/his.13866. Epub 2019 Jun 25. Histopathology. 2019. PMID: 30908700 Review.
Cited by
-
Comparative transcriptomics of primary cells in vertebrates.Genome Res. 2020 Jul;30(7):951-961. doi: 10.1101/gr.255679.119. Epub 2020 Jul 27. Genome Res. 2020. PMID: 32718981 Free PMC article.
-
Nonsense-Mediated Decay Targeted RNA (ntRNA): Proposal of a ntRNA-miRNA-lncRNA Triple Regulatory Network Usable as Biomarker of Prognostic Risk in Patients with Kidney Cancer.Genes (Basel). 2022 Sep 15;13(9):1656. doi: 10.3390/genes13091656. Genes (Basel). 2022. PMID: 36140823 Free PMC article.
-
MicroRNA-490-3p suppresses hepatocellular carcinoma cell proliferation and migration by targeting the aurora kinase A gene (AURKA).Arch Med Sci. 2019 Dec 31;16(2):395-406. doi: 10.5114/aoms.2019.91351. eCollection 2020. Arch Med Sci. 2019. PMID: 32190151 Free PMC article.
-
Therapeutic Targeting of TFE3/IRS-1/PI3K/mTOR Axis in Translocation Renal Cell Carcinoma.Clin Cancer Res. 2018 Dec 1;24(23):5977-5989. doi: 10.1158/1078-0432.CCR-18-0269. Epub 2018 Jul 30. Clin Cancer Res. 2018. PMID: 30061365 Free PMC article.
-
The suitability of NONO-TFE3 dual-fusion FISH assay as a diagnostic tool for NONO-TFE3 renal cell carcinoma.Sci Rep. 2020 Oct 1;10(1):16361. doi: 10.1038/s41598-020-73309-4. Sci Rep. 2020. PMID: 33004995 Free PMC article.
References
-
- Argani P, Antonescu CR, Couturier J, Fournet J-C, Sciot R, Debiec-Rychter M, et al. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11. 2;q21) Am J Surg Pathol. 2002;26:1553–66. - PubMed
-
- Sukov WR, Hodge JC, Lohse CM, Leibovich BC, Thompson RH, Pearce KE, et al. TFE3 rearrangements in adult renal cell carcinoma: clinical and pathologic features with outcome in a large series of consecutively treated patients. Am J Surg Pathol. 2012;36:663–70. doi: 10.1097/PAS.0b013e31824dd972. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

