Whole-Exome Sequencing Identifies Biallelic IDH3A Variants as a Cause of Retinitis Pigmentosa Accompanied by Pseudocoloboma
- PMID: 28412069
- PMCID: PMC5868413
- DOI: 10.1016/j.ophtha.2017.03.010
Whole-Exome Sequencing Identifies Biallelic IDH3A Variants as a Cause of Retinitis Pigmentosa Accompanied by Pseudocoloboma
Abstract
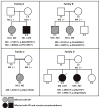
Purpose: To identify the genetic cause of and describe the phenotype in 4 families with autosomal recessive retinitis pigmentosa (arRP) that can be associated with pseudocoloboma.
Design: Case series.
Participants: Seven patients from 4 unrelated families with arRP, among whom 3 patients had bilateral early-onset macular pseudocoloboma.
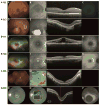
Methods: We performed homozygosity mapping and whole-exome sequencing in 5 probands and 2 unaffected family members from 4 unrelated families. Subsequently, Sanger sequencing and segregation analysis were performed in additional family members. We reviewed the medical history of individuals carrying IDH3A variants and performed additional ophthalmic examinations, including full-field electroretinography, fundus photography, fundus autofluorescence imaging, and optical coherence tomography.
Main outcome measures: IDH3A variants, age at diagnosis, visual acuity, fundus appearance, visual field, and full-field electroretinography, fundus autofluorescence, and optical coherence tomography findings.
Results: We identified 7 different variants in IDH3A in 4 unrelated families, that is, 5 missense, 1 nonsense, and 1 frameshift variant. All participants showed symptoms early in life, ranging from night blindness to decreased visual acuity, and were diagnosed between the ages of 1 and 11 years. Four participants with biallelic IDH3A variants displayed a typical arRP phenotype and 3 participants were diagnosed with arRP and pseudocoloboma of the macula.
Conclusions: IDH3A variants were identified as a novel cause of typical arRP in some individuals associated with macular pseudocoloboma. We observed both phenotypes in 2 siblings carrying the same compound heterozygous variants, which could be explained by variable disease expression and warrants caution when making assertions about genotype-phenotype correlations.
Copyright © 2017 American Academy of Ophthalmology. All rights reserved.
Figures

References
-
- Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;(233):1–34. - PubMed
-
- Rosner M, Hefetz L, Abraham FA. The prevalence of retinitis pigmentosa and congenital stationary night blindness in Israel. Am J Ophthalmol. 1993;116(3):373–4. - PubMed
-
- Grondahl J. Estimation of prognosis and prevalence of retinitis pigmentosa and Ushersyndrome in Norway. Clin Genet. 1987;31(4):255–64. - PubMed
-
- Bunker CH, Berson EL, Bromley WC, et al. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97(3):357–65. - PubMed
-
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–809. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

