Spop regulates Gli3 activity and Shh signaling in dorsoventral patterning of the mouse spinal cord
- PMID: 28412462
- PMCID: PMC5638672
- DOI: 10.1016/j.ydbio.2017.04.002
Spop regulates Gli3 activity and Shh signaling in dorsoventral patterning of the mouse spinal cord
Abstract
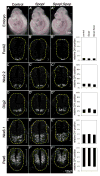
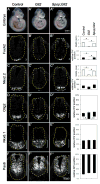
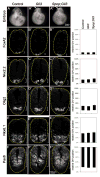
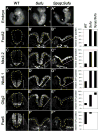
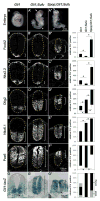
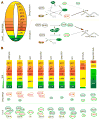
Sonic Hedgehog (Shh) signaling regulates the patterning of ventral spinal cord through the effector Gli family of transcription factors. Previous in vitro studies showed that an E3 ubiquitin ligase containing Speckle-type POZ protein (Spop) targets Gli2 and Gli3 for ubiquitination and degradation, but the role of Spop in Shh signaling and mammalian spinal cord patterning remains unknown. Here, we show that loss of Spop does not alter spinal cord patterning, but it suppresses the loss of floor plate and V3 interneuron phenotype of Gli2 mutants, suggesting a negative role of Spop in Gli3 activator activity, Shh signaling and the specification of ventral cell fates in the spinal cord. This correlates with a moderate but significant increase in the level of Gli3 protein in the Spop mutant spinal cords. Furthermore, loss of Spop restores the maximal Shh pathway activation and ventral cell fate specification in the Gli1;Sufu double mutant spinal cord. Finally, we show that loss of Spop-like does not change the spinal cord patterning in either wild type or Spop mutants, suggesting that it does not compensate for the loss of Spop in Shh signaling and spinal cord patterning. Therefore, our results demonstrate a negative role of Spop in the level and activity of Gli3, Shh signaling and ventral spinal cord patterning.
Keywords: Gli1; Gli2; Neural patterning; Spopl; Sufu; Ubiquitin ligase.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3.Dev Cell. 2004 Jan;6(1):103-15. doi: 10.1016/s1534-5807(03)00394-0. Dev Cell. 2004. PMID: 14723851
-
The amino-terminal region of Gli3 antagonizes the Shh response and acts in dorsoventral fate specification in the developing spinal cord.Dev Biol. 2003 May 15;257(2):343-55. doi: 10.1016/s0012-1606(03)00065-4. Dev Biol. 2003. PMID: 12729563
-
Combinatorial Gli gene function in floor plate and neuronal inductions by Sonic hedgehog.Development. 1998 Jun;125(12):2203-12. doi: 10.1242/dev.125.12.2203. Development. 1998. PMID: 9584120
-
Gli proteins and the control of spinal-cord patterning.EMBO Rep. 2003 Aug;4(8):761-5. doi: 10.1038/sj.embor.embor896. EMBO Rep. 2003. PMID: 12897799 Free PMC article. Review.
-
The emergent design of the neural tube: prepattern, SHH morphogen and GLI code.Curr Opin Genet Dev. 2003 Oct;13(5):513-21. doi: 10.1016/j.gde.2003.08.005. Curr Opin Genet Dev. 2003. PMID: 14550418 Review.
Cited by
-
Challenges and opportunities for the diverse substrates of SPOP E3 ubiquitin ligase in cancer.Theranostics. 2025 May 8;15(13):6111-6145. doi: 10.7150/thno.113356. eCollection 2025. Theranostics. 2025. PMID: 40521202 Free PMC article. Review.
-
SPOP in Cancer: Phenomena, Mechanisms and Its Role in Therapeutic Implications.Genes (Basel). 2022 Nov 7;13(11):2051. doi: 10.3390/genes13112051. Genes (Basel). 2022. PMID: 36360288 Free PMC article. Review.
-
Nabais Sa-de Vries syndrome in a Chinese infant associated with a novel SPOP mutation: A clinical study and genetic report.Mol Genet Genomic Med. 2022 Dec;10(12):e2075. doi: 10.1002/mgg3.2075. Epub 2022 Oct 19. Mol Genet Genomic Med. 2022. PMID: 36259278 Free PMC article.
-
SPOP attenuates migration and invasion of choriocarcinoma cells by promoting DHX9 degradation.Am J Cancer Res. 2020 Aug 1;10(8):2428-2445. eCollection 2020. Am J Cancer Res. 2020. PMID: 32905556 Free PMC article.
-
TMEM132A regulates mouse hindgut morphogenesis and caudal development.Development. 2023 Jul 15;150(14):dev201630. doi: 10.1242/dev.201630. Epub 2023 Jul 13. Development. 2023. PMID: 37390294 Free PMC article.
References
-
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–61. - PubMed
-
- Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–72. - PubMed
-
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–15. - PubMed
-
- Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

