Fatty acid metabolism in breast cancer subtypes
- PMID: 28412757
- PMCID: PMC5438746
- DOI: 10.18632/oncotarget.15494
Fatty acid metabolism in breast cancer subtypes
Abstract
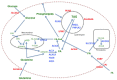
Dysregulation of fatty acid metabolism is recognized as a component of malignant transformation in many different cancers, including breast; yet the potential for targeting this pathway for prevention and/or treatment of cancer remains unrealized. Evidence indicates that proteins involved in both synthesis and oxidation of fatty acids play a pivotal role in the proliferation, migration and invasion of breast cancer cells. The following essay summarizes data implicating specific fatty acid metabolic enzymes in the genesis and progression of breast cancer, and further categorizes the relevance of specific metabolic pathways to individual intrinsic molecular subtypes of breast cancer. Based on mRNA expression data, the less aggressive luminal subtypes appear to rely on a balance between de novo fatty acid synthesis and oxidation as sources for both biomass and energy requirements, while basal-like, receptor negative subtypes overexpress genes involved in the utilization of exogenous fatty acids. With these differences in mind, treatments may need to be tailored to individual subtypes.
Keywords: breast cancer; fatty acid metabolism; molecular subtype.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
References
-
- Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. Journal of Clinical Oncology. 2009;27:1160–7. doi: 10.1200/jco.2008.18.1370. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

