Automated Wormscan
- PMID: 28413617
- PMCID: PMC5365223
- DOI: 10.12688/f1000research.10767.3
Automated Wormscan
Abstract
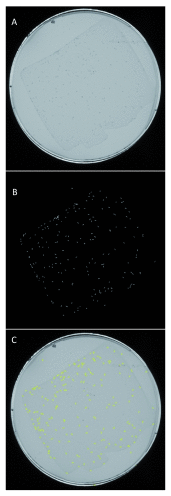
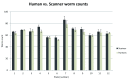
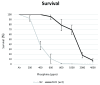
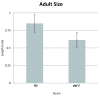
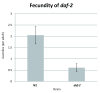
There has been a recent surge of interest in computer-aided rapid data acquisition to increase the potential throughput and reduce the labour costs of large scale Caenorhabditis elegans studies. We present Automated WormScan, a low-cost, high-throughput automated system using commercial photo scanners, which is extremely easy to implement and use, capable of scoring tens of thousands of organisms per hour with minimal operator input, and is scalable. The method does not rely on software training for image recognition, but uses the generation of difference images from sequential scans to identify moving objects. This approach results in robust identification of worms with little computational demand. We demonstrate the utility of the system by conducting toxicity, growth and fecundity assays, which demonstrate the consistency of our automated system, the quality of the data relative to manual scoring methods and congruity with previously published results.
Keywords: Caenorhabditis elegans; WormScan; phosphine; software; toxicology.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

