L1000CDS2: LINCS L1000 characteristic direction signatures search engine
- PMID: 28413689
- PMCID: PMC5389891
- DOI: 10.1038/npjsba.2016.15
L1000CDS2: LINCS L1000 characteristic direction signatures search engine
Abstract
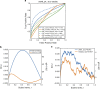
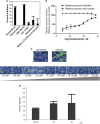
The library of integrated network-based cellular signatures (LINCS) L1000 data set currently comprises of over a million gene expression profiles of chemically perturbed human cell lines. Through unique several intrinsic and extrinsic benchmarking schemes, we demonstrate that processing the L1000 data with the characteristic direction (CD) method significantly improves signal to noise compared with the MODZ method currently used to compute L1000 signatures. The CD processed L1000 signatures are served through a state-of-the-art web-based search engine application called L1000CDS2. The L1000CDS2 search engine provides prioritization of thousands of small-molecule signatures, and their pairwise combinations, predicted to either mimic or reverse an input gene expression signature using two methods. The L1000CDS2 search engine also predicts drug targets for all the small molecules profiled by the L1000 assay that we processed. Targets are predicted by computing the cosine similarity between the L1000 small-molecule signatures and a large collection of signatures extracted from the gene expression omnibus (GEO) for single-gene perturbations in mammalian cells. We applied L1000CDS2 to prioritize small molecules that are predicted to reverse expression in 670 disease signatures also extracted from GEO, and prioritized small molecules that can mimic expression of 22 endogenous ligand signatures profiled by the L1000 assay. As a case study, to further demonstrate the utility of L1000CDS2, we collected expression signatures from human cells infected with Ebola virus at 30, 60 and 120 min. Querying these signatures with L1000CDS2 we identified kenpaullone, a GSK3B/CDK2 inhibitor that we show, in subsequent experiments, has a dose-dependent efficacy in inhibiting Ebola infection in vitro without causing cellular toxicity in human cell lines. In summary, the L1000CDS2 tool can be applied in many biological and biomedical settings, while improving the extraction of knowledge from the LINCS L1000 resource.
Conflict of interest statement
COMPETING INTERESTS The authors declare no conflict of interest.
Figures



Similar articles
-
Integrating Differential Gene Expression Analysis with Perturbagen-Response Signatures May Identify Novel Therapies for Thyroid-Associated Orbitopathy.Transl Vis Sci Technol. 2020 Aug 25;9(9):39. doi: 10.1167/tvst.9.9.39. eCollection 2020 Aug. Transl Vis Sci Technol. 2020. PMID: 32908802 Free PMC article.
-
LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures.Nucleic Acids Res. 2014 Jul;42(Web Server issue):W449-60. doi: 10.1093/nar/gku476. Epub 2014 Jun 6. Nucleic Acids Res. 2014. PMID: 24906883 Free PMC article.
-
Compound signature detection on LINCS L1000 big data.Mol Biosyst. 2015 Mar;11(3):714-22. doi: 10.1039/c4mb00677a. Epub 2015 Jan 22. Mol Biosyst. 2015. PMID: 25609570 Free PMC article.
-
The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations.Cell Syst. 2018 Jan 24;6(1):13-24. doi: 10.1016/j.cels.2017.11.001. Epub 2017 Nov 29. Cell Syst. 2018. PMID: 29199020 Free PMC article. Review.
-
[Development of antituberculous drugs: current status and future prospects].Kekkaku. 2006 Dec;81(12):753-74. Kekkaku. 2006. PMID: 17240921 Review. Japanese.
Cited by
-
Coupled immune stratification and identification of therapeutic candidates in patients with lung adenocarcinoma.Aging (Albany NY). 2020 Aug 27;12(16):16514-16538. doi: 10.18632/aging.103775. Epub 2020 Aug 27. Aging (Albany NY). 2020. PMID: 32855362 Free PMC article.
-
signatureSearch: environment for gene expression signature searching and functional interpretation.Nucleic Acids Res. 2020 Dec 2;48(21):e124. doi: 10.1093/nar/gkaa878. Nucleic Acids Res. 2020. PMID: 33068417 Free PMC article.
-
Discovery of preventive drugs for cisplatin-induced acute kidney injury using big data analysis.Clin Transl Sci. 2022 Jul;15(7):1664-1675. doi: 10.1111/cts.13282. Epub 2022 Apr 30. Clin Transl Sci. 2022. PMID: 35445533 Free PMC article.
-
Drug repurposing based on differentially expressed genes suggests drug combinations with possible synergistic effects in treatment of lung adenocarcinoma.Cancer Biol Ther. 2023 Dec 31;24(1):2253586. doi: 10.1080/15384047.2023.2253586. Cancer Biol Ther. 2023. PMID: 37710391 Free PMC article.
-
A Bayesian approach to accurate and robust signature detection on LINCS L1000 data.Bioinformatics. 2020 May 1;36(9):2787-2795. doi: 10.1093/bioinformatics/btaa064. Bioinformatics. 2020. PMID: 32003771 Free PMC article.
References
-
- Stegmaier, K. et al. Gene expression–based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat. Genet. 36, 257–263 (2004). - PubMed
-
- Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). - PubMed
-
- Campillos, M., Kuhn, M., Gavin, A.-C., Jensen, L. J. & Bork, P. Drug target identification using side-effect similarity. Science 321, 263–266 (2008). - PubMed
-
- Smyth, G. K. in Bioinformatics and Computational Biology Solutions Using R and Bioconductor 397–420 (Springer, 2005).
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

